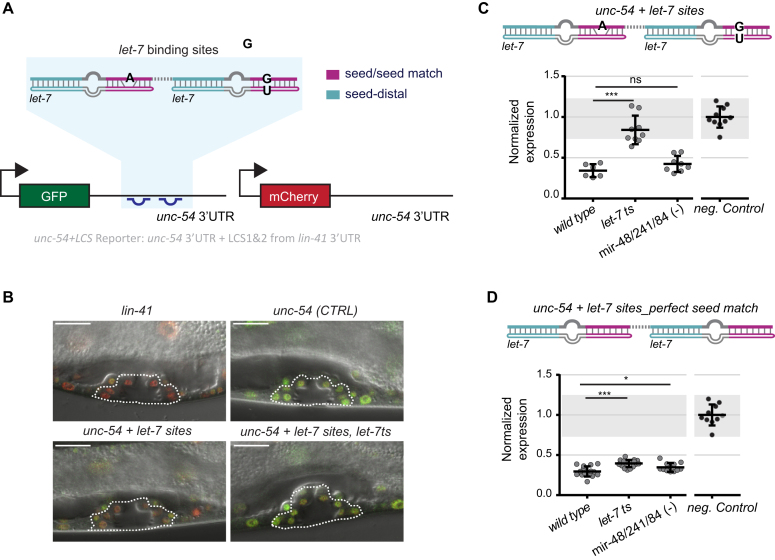
Figure 2.
Redundant activity of the let-7 family in the presence of a perfect seed match. (A) Schematic of the reporters used to monitor miRNA activity in vivo. The depicted GFP transgene unc-54 + let-7 sites reporter contains 111 nucleotides of the lin-41 3′UTR (shaded in blue), which harbor the two let-7 binding sites and the 27 nt-long intervening sequence, grafted into the heterologous, unregulated unc-54 3′UTR. Worms also contain a red mCherry reporter for normalization. Transcription of the single-copy integrated reporters from the ubiquitously active dpy-30 promoter is constitutive. miRNA site legend: magenta = seed/seed match; cyan = let-7 seed-distal binding. (B) Representative confocal images of the vulvae of animals carrying the red mCherry reporter (for normalization) and GFP reporters with the indicated 3′UTRs. These are ‘lin-41 3′UTR full-length’, ‘unc-54’ (CTRL, unregulated) and ‘unc-54 + let-7 sites’ in wild-type and ‘unc-54 + let-7 sites’ in the let-7ts background. Images are merged GFP, mCherry and DIC channels. Red color indicates a greater, and green color a lesser degree of reporter repression. Dashed lines outline the vulvae of the animals, which confirm appropriate late Larval stage 4 (L4). Scale bars 15 μm. (C, D) Quantification of (C) ‘unc-54 + let-7 sites’ reporter, (D) ‘unc-54 + let-7 sites_perfect seed match’ reporter. Each dot represents the average of the GFP signal intensity, obtained by confocal imaging, divided by the mCherry intensity for a single animal per condition. 10–12 vulva cells were quantified per worm. Mean values are normalized to the average value of the GFP/mCherry ratio of the negative control unc-54 3′UTR reporter, which is not silenced. Horizontal line and error bars indicate mean values per condition ± SD. *P < 0.05 and ***P < 0.001, two-tailed unpaired t-test. For reference, data obtained for the unc-54, Neg.Control reporter are replotted in panel D; gray shading is bounded by the min-max values of this control.