Figure 4.
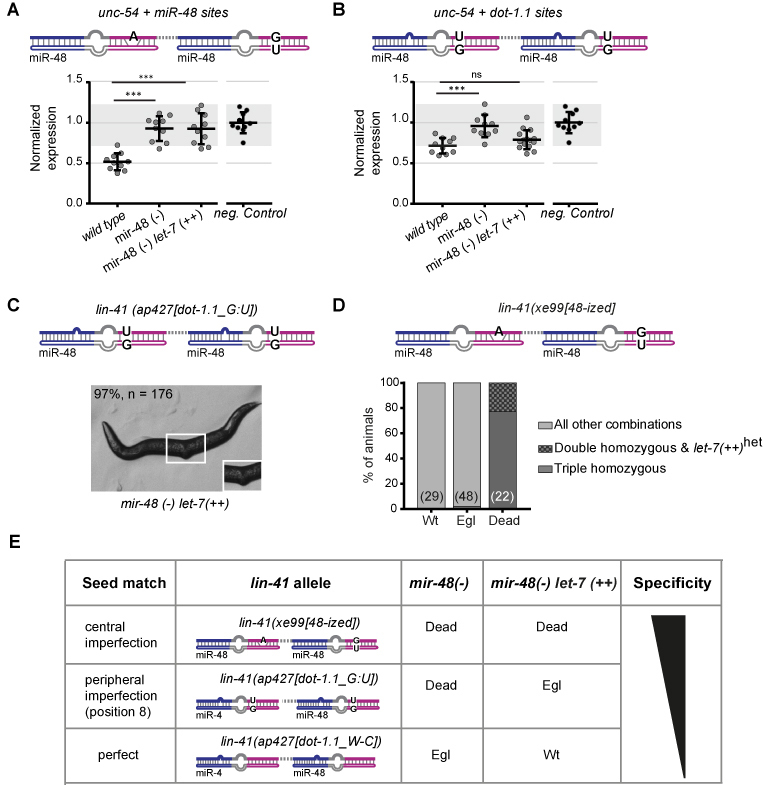
Robust miRNA specificity relies on imperfect seed matches. (A, B) Reporter quantification as in Figure 2, from which the negative control data are also replotted for reference. (A) ‘unc-54 + miR-48 sites’ reporter and (B) ‘unc-54 + dot-1.1 sites’ reporter are assayed in worms of the indicated genotypes. Horizontal line and error bars indicate mean values per condition ± SD, *P < 0.05 and ***P < 0.001, two-tailed unpaired t-test. (C) Representative image of a viable lin-41(ap427[dot-1.1_G:U]), mir-48(-) let-7(++) animal. (D) Progeny (n = 99) derived from a cross of lin-41(xe99[48-ized]) with mir-48(–) let-7(++) animals were categorized by phenotype and genotyped to determine the viability of lin-41(xe99[48-ized]); mir-48(-) let-7(++) ‘triple homozygous’ mutant animals. (E) Summary of the effect that different site architectures and miRNA abundance have on silencing lin-41 alleles ‘recoded’ towards miR-48. mir-48(–): mir-48(n4097)V; unc-54(CTRL): wild-type unc-54 3′UTR; let-7(++): let-7 over-expression allele (MosSCI, V).
