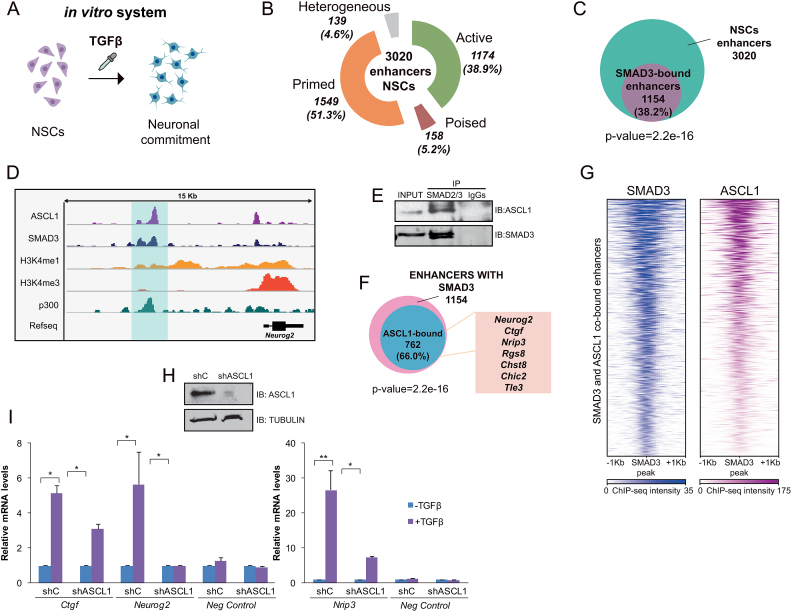
Figure 1.
Epigenomic identification of neural enhancers. (A) Schematic view of the model used to study neural enhancer activation upon the TGFβ differentiation signal. (B) Number and type of identified enhancers using ChIP-seq data for p300, H3K4me1, H3K4me3, H3K27ac and H3K27me3. The percentage related to the total number of enhancers is displayed in the graphic. (C) Venn diagram showing the number of neural enhancers bound by SMAD3 in NSCs treated with TGFβ for 30 min. P-value is the result of an equal proportions test performed between SMAD3-bound enhancers and a random set. (D) IGV capture showing the chromatin landscape around the Neurog2 gene. Neurog2(-6) enhancer is highlighted in blue. Tracks display ChIP-seq in NSCs treated for 30 min with TGFβ (SMAD3) or untreated cells (ASCL1, p300, H3K4me1 and H3K4me3). (E) Endogenous SMAD2/3 was immunoprecipitated from NSCs and the presence of SMAD3 and ASCL1 in the immunopellet was determined by immunoblot with the antibodies indicated on the right part of the figure. Figure is representative of at least two biological independent experiments. (F) Venn diagram showing the number of neural SMAD3-bound enhancers upon TGFβ treatment for 30 min that contain ASCL1 TF in untreated NSCs. The names of some genes putatively regulated by these enhancers are indicated. (G) Heatmap representation of SMAD3 (left) and ASCL1 (right) binding on the SMAD3 and ASCL1 co-occupied enhancers. Scales indicate ChIP-seq intensities. (H) NSCs were infected with lentivirus expressing shRNA control (shC) or shRNA specific for ASCL1 (shASCL1) cloned into pLKO vector. Forty eight hours later, total protein extracts were prepared and the ASCL1 and TUBULIN levels were determined by immunoblot. (I) shC and shASCL1 cells were treated with TGFβ for 6 h (3 h for Ctgf) and mRNA levels of TGFβ-responsive genes associated to the enhancers bound by SMAD3 and ASCL1 were analyzed by qPCR. Data were normalized to Rps23 housekeeping gene and figure shows values relative to shC samples. Ccne3 gene, a non TGFβ-responsive gene was used as a control. Results are the mean of three biological independent experiments. Error bars indicate SD. *P < 0.05; **P < 0.01 (Student's t-test).