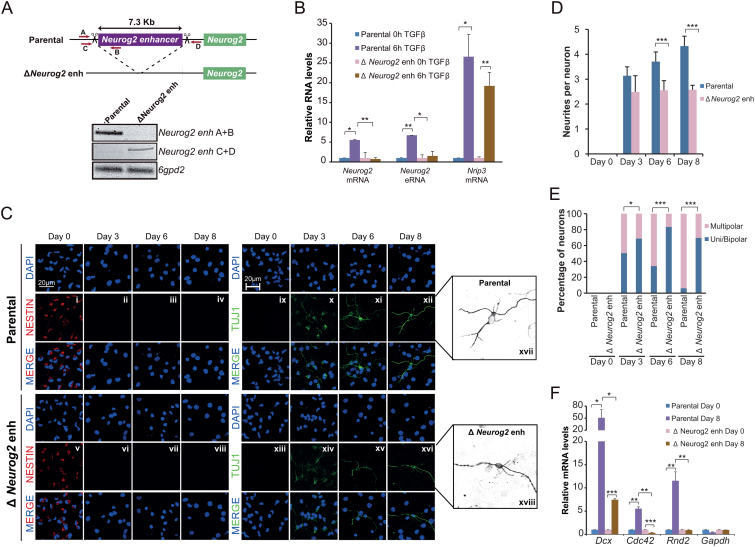
Figure 5.
TGFβ-responsive enhancers are essential for neuronal differentiation. (A) Schematic representation of the CRISPR/Cas9 experimental approach used to delete Neurog2(–6) enhancer in NSCs. Two gRNAs flanking Neurog2(-6) enhancer region were used to create the deletion. Red arrows represent primers to test the deletion. PCR using Neurog2 deletion and 6gpd2 pairs of primers is shown in the top of the figure in parental and ΔNeurog2 enh NSC lines. (B) Parental and ΔNeurog2 enh cell lines were treated with TGFβ for 6 h. Total RNA was prepared and the expression levels of Neurog2 mRNA or eRNA were determined by qPCR. mRNA level of Nrip3 was used as a TGFβ response control. Transcription values were normalized to the housekeeping gene Rps23, and figure shows values relative to time 0 h. Errors bars represent SD. Results are representatives of two biological independent experiments. *P < 0.05; **P < 0.01 (Student's t-test). (C–E) Parental and ΔNeurog2 enh cell lines were cultured in differentiating medium. After 3, 6 or 8 days cells were fixed and stained with NESTIN, TUJ1 antibody and DAPI (C). Number of neurites per cell (D) and the percentage of uni/bipolar or multipolar pseudoneurons (E) were quantified by direct counting of 10 randomly selected fields. Data show mean of n = 60 cells. Error bars indicate SD. *P < 0.05; ***P < 0.001 (Student's t-test). (F) Parental and ΔNeurog2 enh cell lines were maintained in differentiating medium for 8 days. Total RNA was purified and the expression levels of the indicated genes were determined by qPCR. Transcription values were normalized to the housekeeping gene Ubc, and figure shows values relative to Day 0 samples. Errors bars represent SD. Results are representatives of two biological independent experiments. *P < 0.05; **P < 0.01; ***P < 0.001 (Student's t-test).