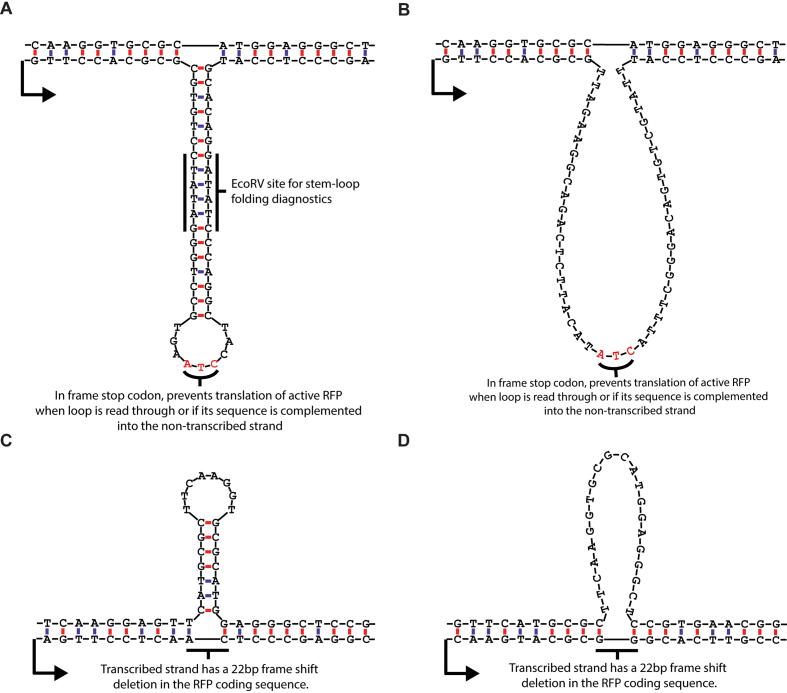
Figure 2.
DNA loop and stem–loop sequences and structures. (A) stem–loop, transcribed strand. The sequence of the extrahelical DNA on the transcribed strand was designed such that the stem–loop contained a restriction site (EcoRV) in the stem with a sufficient length of dsDNA (6 base pairs) on either side of the restriction site for recognition and cutting. This was done for diagnostic purposes to confirm that the stem loop folded as expected (Supplementary Figure S2). (B) Unpaired loop, transcribed strand. The unpaired loop on the transcribed strand was matched in length to the stem–loop structure. Both sequences contained in-frame stop codons for cell culture experiments, where removal of the loop or stem–loop from the transcribed strand would lead to RFP activation, but retention or expansion of the sequence would produce a truncated, non-functional RFP. (C) Stem–loop, non-transcribed strand. The sequences of the extrahelical DNA on the non-transcribed strand were limited by the native RFP coding sequence. The length of 22 nucleotides was chosen because there was a sequence in the RFP coding region that would form a stem loop with a 7 base-pair stem and a 9 nucleotide loop if the corresponding sequence on the transcribed strand were missing. The 22 nucleotide deletion on the transcribed strand also caused a frame-shift mutation that produced a non-functional protein product in cell culture experiments. (D) Unpaired loop, non-transcribed strand. Similar to the stem–loop on the non-transcribed strand, this is a native sequence in the RFP coding sequence that was not predicted to form significant internal base-pairing. The transcribed strand also has a 22 nucleotide frame-shift deletion in the RFP coding sequence that knocks out fluorescence.