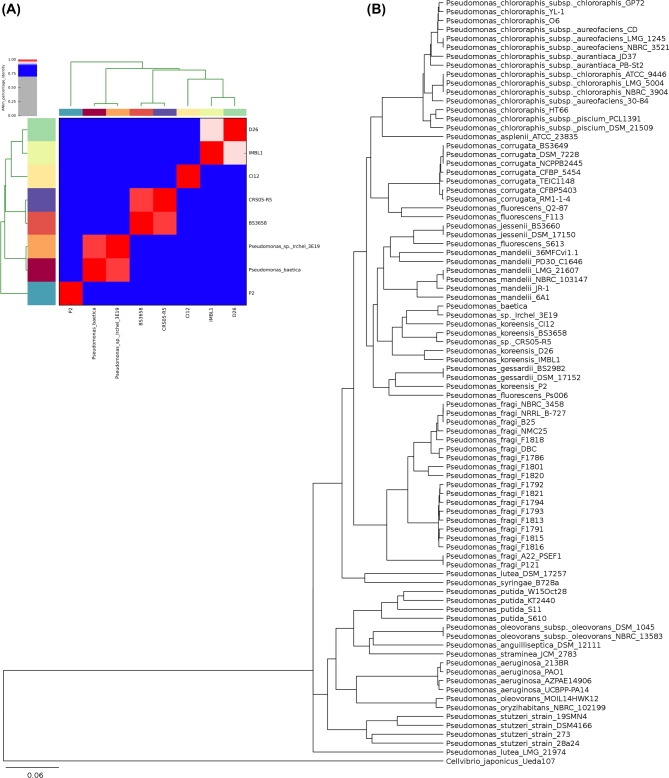
Figure 1.
Placement of novel baetica genome in the genus Pseudomonas based upon ANI, and shared k-mers A) ANI heatmap generated the Python3 module pyani. The sequenced baetica genome, Pseudomonas_sp._Irchel_3E19 and reference genomes from the closely related koreensis subclade were subject to ANI analysis. B) A Mash-based tree generated from reference genomes (= 86) from 19 species clades, comprising the entire genus Pseudomonas. This tree was generated based upon the Jaccard index, calculated from shared k-mers. Cellvibrio japonicum Ueda107 was used as outgroup.