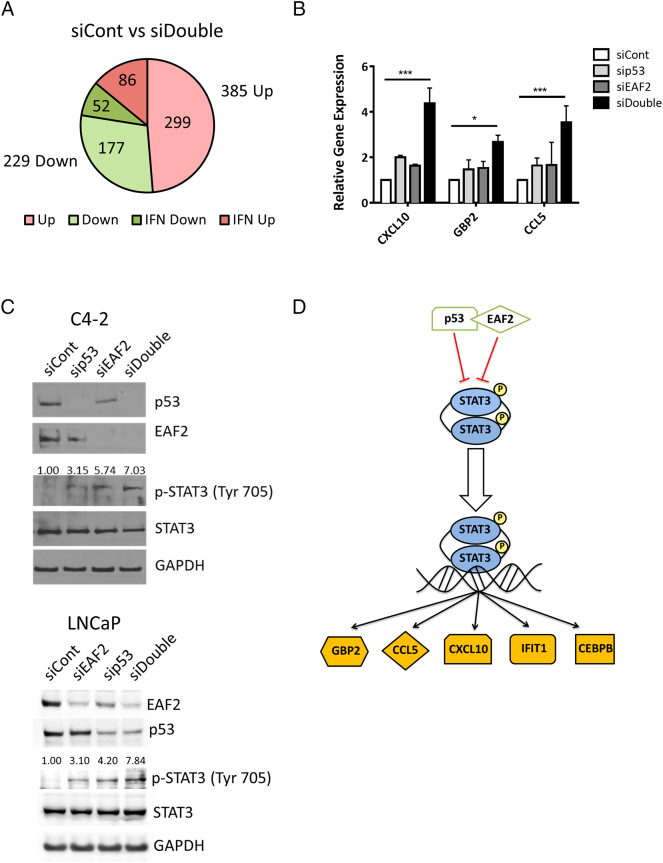
Figure 2.
(A) RNA-seq identification of 614 altered genes in C4-2 cells treated with siRNA control (siCont) compared to concurrent siEAF2 and sip53 (siDouble). Of the 385 upregulated genes (Up), 86 were identified as interferon-regulated (IFN) genes. Of the 229 downregulated genes (Down), 52 were identified as IFN-regulated genes. (B) qPCR verification of several STAT3 target genes identified by RNA-seq as upregulated in C4-2 p53 and EAF2 double-knockdown cells expressed as mean ± S.D. relative to control (siCont). (C) Western immunoblotting of STAT3 phosphorylation (Tyr 705) in C4-2 cells (top panel) and LNCaP cells (bottom panel) with knockdown of p53 and/or EAF2. GAPDH served as loading control. (D). Graphical depiction of potential EAF2 and p53 interaction in the STAT3 pathway. Gold molecules represent genes identified by RNA-Seq as upregulated in siDouble C4-2 cells.