Figure 1. SPaDES framework for high-resolution membrane protein modeling.
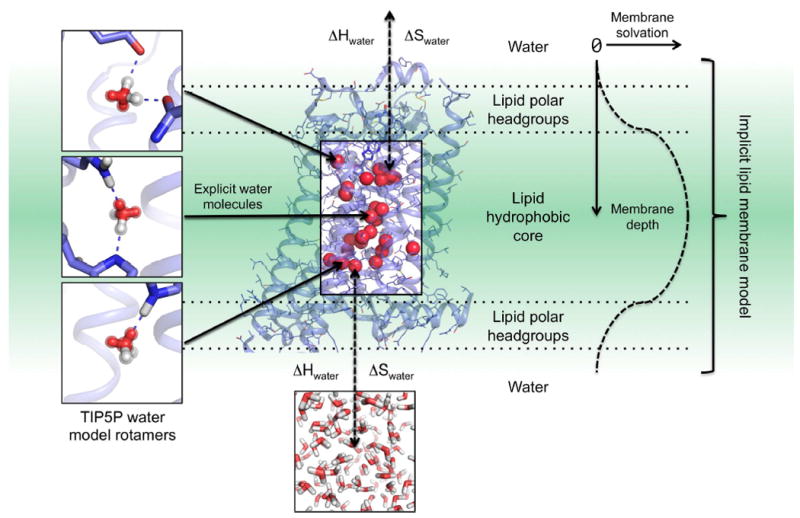
Transmembrane protein structures are modeled with a hybrid solvent representation where buried water molecules are modeled explicitly in the protein core (middle box) and the protein environment (lipid, water, and ion molecules) is modeled implicitly (green background). Explicit buried solvent molecules are modeled with Tip5p water rotamers anchored by hydrogen bonds to unsatisfied protein polar atoms in either a bridging configuration (top and middle left panels) or a single bond (bottom left panel). Water molecules are predicted to stay within the interior of the protein when the enthalpic (ΔH) and entropic (ΔS) cost of removing them from the solvent (ΔH and ΔS) is lower than the binding energy with the protein (dotted arrows). Otherwise, water molecules move back to the bulk solvent (bottom panel). Protein interactions with the implicit lipid membrane are described by the RosettaMembrane energy function, which accounts for the membrane depth and solvation (right diagrams and labels).
