Figure 2. Accurate de novo prediction of protein-bound water molecule positions.
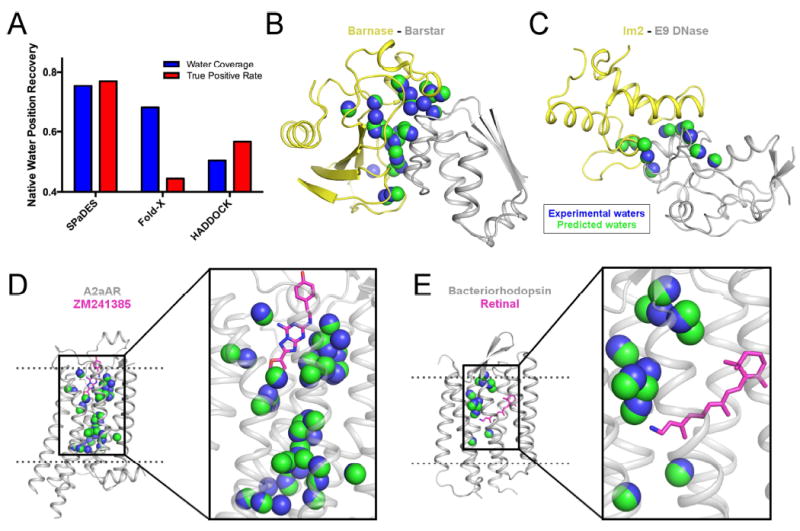
(A) Recovery of experimentally-observed water molecules (blue) and true positive rates of predicted water molecules (red) are shown with SPaDES, Fold-X with explicit water molecule modeling, and HADDOCK refinement with a 2.0 Å distance cutoff critera between predicted and experimentally-resolved waters. Higher fractional rates, ranging from zero to one, indicate increased accuracy of the predictions. (B-E) Examples of predicted water positions are shown for two protein-protein complexes and two integral ligand-bound membrane proteins: (B) barnase in complex with barstar, (C) colicin E2 immunity protein in complex with colicin E9 DNase, (D) A2a adenosine receptor bound to the ZM241385 ligand, and (E) bacteriorhodopsin bound to the retinal ligand. Blue spheres: water molecules observed in protein X-ray structures (“experimental waters”). Green spheres: de novo predicted water molecules (“predicted waters”). Ligands are shown in magenta sticks. See also Figure S2, Table S1.
