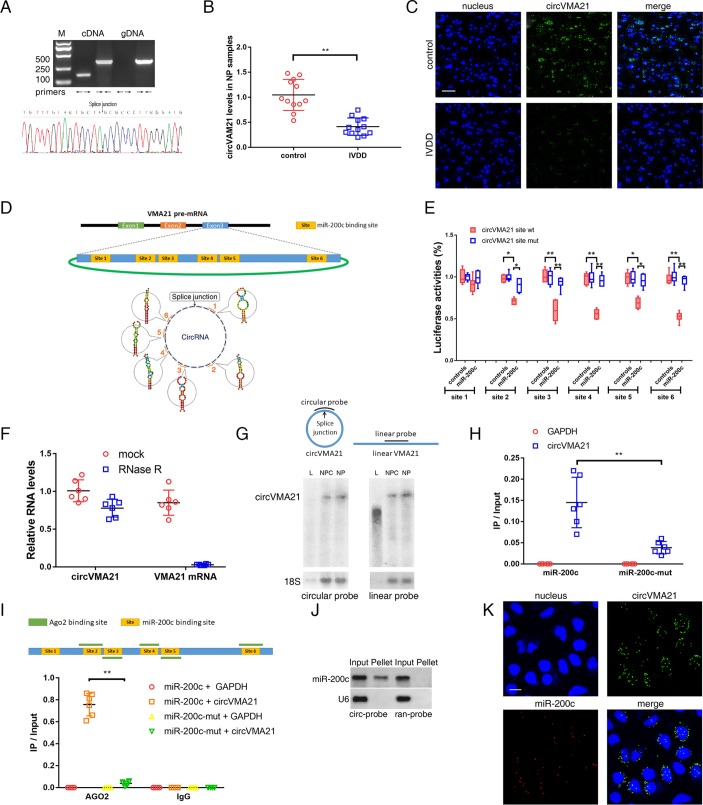
Figure 3.
circVMA21 acted as a sponge of miR-200c. (A) Agarose gel electrophoresis found that divergent primers (←→) amplified circVMA21 in complementary DNA (cDNA) but not genomic DNA (gDNA) (upper). The amplified product of specific divergent primers was confirmed in accordance with the sequence of circVMA21 by sequencing (lower). (B) qRT-PCR analysis detected circVMA21 levels in the NP samples from patients with or without IVDD. n=12; **P<0.01. (C) The expression of circVMA21 was detected in the NP samples from patients with or without IVDD by RNA fluorescence in situ hybridisation (FISH). circVMA21 probe was labelled with Alexa 488. Nuclei were stained with 4,6-diamidino-2-phenylindole (DAPI). Scale bar=50 µm. (D) circVMA21 is transcribed from the third exon of the VMA21 gene and contains six putative binding sites complementary to miR-200c. (E) NPCs were transfected with miR-200c and luciferase constructs of circVMA21 containing wild-type putative miR-200c binding sites (circVMA21 site wt) or mutated sites (circVMA21 site mut). n=6; *P<0.05, **P<0.01. (F) qRT-PCR analysis for the abundance of circVMA21 and VMA21 mRNA in NPCs with or without RNase R treatment. The amounts were normalised to the value of circVMA21 measured in the mock treatment. n=6. (G) Northern blot analysis showed that linear VMA21 was detectable by a linear but not circular probe. L, linear VMA21 transcribed in vitro; NPC, total RNAs extracted from NPCs; NP, total RNAs extracted from NP tissue samples; circular probe, probe within splice site; linear probe, head-to-tail probe. (H) The biotinylated miR-200c or its mutant (miR-200c-mut) was transfected into NPCs. The RNA levels of circVMA21 and GAPDH were quantified by qRT-PCR analysis, and the relative ratios of immunoprecipitate (IP) to input were plotted. **P<0.01. (I) CLIP sequence revealed five AGO2-bound regions overlapped with the binding sites of miR-200c within the circVMA21 sequence (upper). AGO2 RNA immunoprecipitation in NPCs transfected with miR-200c or its mutant. The levels of circVMA21 and GAPDH were quantified by qRT-PCR analysis, and the relative ratios of IP to input were plotted. **P<0.01 (lower). (J) miR-200c was pulled down by the circular probe for circVMA21 (circ-probe) but not random probe (ran-probe). The levels of miR-200c were detected by northern blot. Input, 20% samples were loaded; Pellet, all samples were loaded. (K) RNA FISH for colocalisation of circVMA21 and miR-200c in cytoplasm of NPCs. circVMA21 and miR-200c probes were labelled with Alexa 488 and Cy-5, respectively. Nuclei were stained with DAPI. Scale bar=10 µm. AGO2, Argonaute 2; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; IVDD, intervertebral disc degeneration; NP, nucleus pulposus; NPC, nucleus pulposus cells; qRT-PCR, quantitative real-time reverse transcription-PCR.