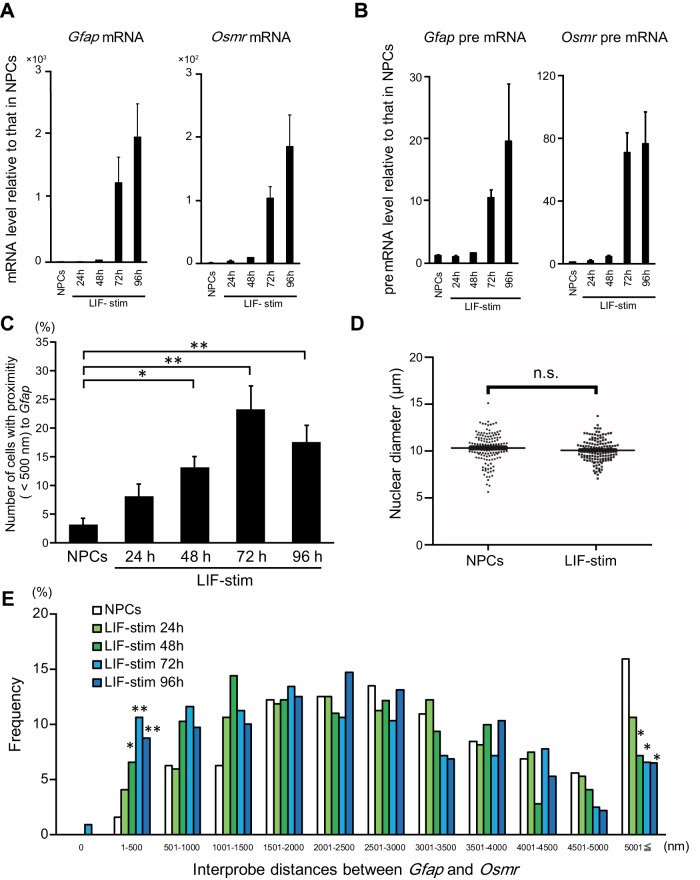
FIGURE 2:
Gfap and Osmr gene cluster in prior to the transcriptional activation of both genes. (A, B) Quantitative RT-PCR was performed on mRNA (A) and pre-mRNA (B) of Gfap and Osmr. The expression level of each gene was determined in NPCs alone and stimulated with LIF for the indicated time (LIF-stim). The results were normalized to Gapdh expression. Each graph represents the mean (±SEM) relative amount (compared with NPCs) of at least three experiments. (C) Clustering frequencies of Gfap and Osmr determined using DNA FISH in NPCs alone and stimulated with LIF for the indicated time (LIF-stim). Each graph represents the mean (±SEM) percentage for the number of cells with clustering alleles of at least three experiments. The Dunnett test was performed. *p < 0.05, **p < 0.01 (n = 53–54). (D) Nuclear diameters of NPCs alone and stimulated with LIF for 96 h (LIF-stim). Nuclear diameters represent the largest diameter of each nucleus stained with DAPI. The Steel test was performed (n = 138). (E) Frequencies of interprobe distances between Gfap and Osmr in NPCs; cells were stimulated with LIF for the indicated time (LIF-stim). The Dunnett test was performed for each interprobe distance. *p < 0.05, **p < 0.01.