Abstract
Formyl peptide receptor-2 (FPR2) is a G protein-coupled receptor belonging to the N-formyl peptide receptor (FPR) family that plays critical roles in peripheral and brain inflammatory responses. FPR2 has been proposed as a target for the development of drugs that could facilitate the resolution of chronic inflammatory reactions by enhancing endogenous anti-inflammation systems. Starting from lead compounds previuosly identified in our laboratories, we designed a new series of ureidopropanamide derivatives with the goal of converting functional activity from agonism to antagonism and to develop new FPR2 antagonists. Although none of the compounds behaved as antagonist, some of the compounds were able to induce receptor desensitization, thus functionally behaving as antagonists. Evaluation of these compounds in an in vitro model of neuroinflammation showed that they reduced reactive oxygen species (ROS) production in mouse microglial N9 cells after stimulation with lipopolysaccharide (LPS). These FPR2 ligands may protect cells from damage due to inflammation-associated oxidative stress.
Keywords: Formyl peptide receptor, ureidopropanamide, neuroinflammation, oxidative stress
Graphical Abstract
Prolonged oxidative stress can trigger cell death and has been implicated in the pathogenesis of many neurodegenerative diseases. We here report on a set of ureidopropanamide derivatives which behave as functional antagonists at Formyl Peptide Receptor 2, a GPCR that plays a critical role in peripheral and central inflammatory responses. Accordingly, the compounds reduce ROS production in LPS-stimulated mouse microglial N9 cells, an in vitro model of neuroinflammation.
Introduction
Inflammation is a complex biological response of the body to harmful stimuli, such as infectious agents, tissue damage, or toxins, and involves several immune and inflammatory cells including neutrophils, macrophages, and mast cells. A successful inflammatory response eliminates invading pathogens, followed by initiation of tissue repair [1]. Similarly, neuroinflammation is a protective mechanism to restore the damaged glial cells and neurons in the central nervous system (CNS). Neuroinflammation is a response that involves all CNS cells, including neurons, microglia and astrocytes, and is mediated by the inflammatory mediators released from these cells [2,3]. Initially, neuroinflammation is a protective response in the brain that initiates the healing process. However, chronic activation of the immune response can lead to excessive release of pro-inflammatory cytokines and neurotoxic mediators, which can lead to neuronal damage and loss [2,3]. Accumulating evidence suggests that chronic neuroinflammation plays an important role in the onset and progression of several neurodegenerative diseases such as Alzheimer’s disease (AD) and Parkinson’s disease (PD) as well as in psychiatric disorders [4,5].
Inflammation is tightly regulated such that pro- and anti-inflammatory mediators operate in a parallel and serial fashion to evoke, at first, the inflammatory response and, then, to ensure resolution of the inflammation [6]. The mechanisms leading to the restoration of homeostasis and resolution of inflammation have been elucidated only recently, and specific pro-resolving mediators, such as lipoxins, resolvins and maresins, have been identified [7,8]. Among the receptors activated by pro-resolving mediators is the formyl peptide receptor 2 (FPR2), a G-protein coupled receptor belonging to the formyl peptide receptor (FPR) family, which also includes the subtypes FPR1 and FPR3 [9]. FPRs were first identified in humans [10] and shortly after in other primates [11] and rodents [12] and are expressed in a variety of tissues and cells, including neutrophils, monocytes/macrophages, and microglia. FPR2 are functionally expressed in glial cells and astrocytes [14–15]. Recently, expression of Fpr1 and Fpr2, murine homologues of human FPR, was also reported in rat neuronal stem cells [17]. FPRs play relevant roles in innate immunity, and their stimulation elicits a cascade of host defense reactions, including chemotaxis, superoxide anion (O2·) generation, and exocytosis [9]. FPR2 interacts with a large number of structurally diverse agonists, such as formylated peptides, amyloidogenic peptides, and prion protein PrP(106-126), which induce pro-inflammatory responses [9]. On the other hand, the N-terminus of the calcium-regulated/phospholipid-binding annexin I and the non-peptide agonists lipoxin A4 (LXA4) and resolvin D1 exert anti-inflammatory and pro-resolving effects, suggesting a biased signaling capacity of FPR2 [9,17].
The involvement of FPR2 in the resolution of inflammation makes it an attractive target for treating a variety of pathologies characterized by chronic inflammation, such as rheumatoid arthritis, asthma, cystic fibrosis, chronic obstructive pulmonary disease, and CNS diseases [18,19]. For example, it has been reported that in vivo administration of LXA4 in rats is able to inhibit microglial activation and to diminish neuroinflammation after spinal cord hemisection [20] and hemorrhage [21]. Likewise, the administration of LXA4 [22] or annexin A1 [23] in animal models of AD is able to stimulate a pro-resolving activation of microglia by reducing the levels of pro-inflammatory cytokines, which results in improved β-amyloid clearance and degradation. Finally, it has been suggested that FPRs are involved in the rapid generation of reactive oxygen species (ROS) from enteric commensal bacteria that can function as second messengers in many signal transduction pathways [24].
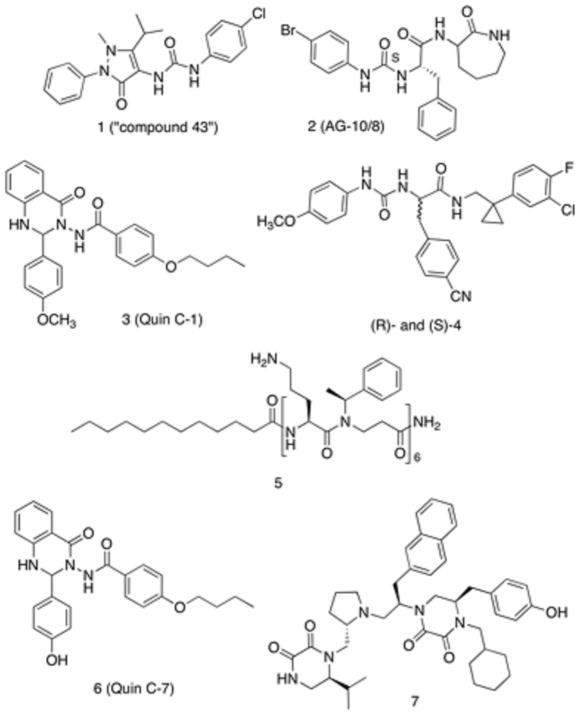
To date, several classes of chemically diverse FPR2 agonists have been reported in the literature [25, 26], such as pyrazolone derivatives like the mixed FPR1/FPR2 agonist 1 (designated also as “compound 43”) [27], N-phenylurea derivatives, such as compound 2 (also named AG-10/8) [28], and quinazolinones derivatives exemplified by the highly specific FPR2 agonists Quin C1 (compound 3) (Figure 1) [29]. Recently, we developed ureidopropanamide derivatives as agonists of human FPR2, exemplified by compounds (R)- and (S)-4 (Figure 1) [30–33]. Regarding FPR2 antagonists, several peptide [9] and peptidomimetic antagonists [34], exemplified by compound 5, have been reported, whereas only a very limited number of small-molecule antagonists have been described so far (Figure 1). Quinazolinone 6, also known as Quin C7, was identified through structure-activity relationship analysis of Quin C1. Compound 6 displays Ki value of 6.7 μM at FPR2 and was characterized as an FPR2 antagonist because it did not activate Ca2+ flux in FPR2 transfected cells and inhibited Ca2+ flux and chemotaxis induced by the FPR2 agonist WKYMVm. Moreover, compound 6 inhibited arachidonic acid-induced ear edema [35]. Another FPR2 antagonist is the pyrrolidine bis-diketopiperazine 7, which was identified by screening combinatorial libraries, and is the most potent non-peptidic FPR2 antagonist identified to date (IC50 = 81 nM) [36]. It is interesting to note that a minor structural change between Quin C1 and Quin C7 (i.e., the removal of the methyl group) converted its functional activity from agonism to antagonism. A docking study performed on different classes of FPR2 ligands suggested that the presence of the hydroxyl group completely changed the binding mode of Quin C7 as compared to that of Quin C1. In particular, Quin C7 lacks interaction with Arg295, which is a critical interaction shared by the majority of FPR2 agonists [37].
Figure 1.
Chemical Structure of FPR2 agonists and antagonists.
Considering the paucity of small-molecule FPR2 antagonists and the inherent limitations of peptides and peptidomimetics as therapeutic agents, we explored the possibility of developing non-peptide antagonists starting from the ureidopropanamide scaffold. We reasoned that insertion of the hydroxyl functionality in the ureidopropanamide scaffold could change the binding mode of the compounds and, therefore, could lead to the interconversion of their functional activity. To this end, we selected compounds (R)- and (S)-4 (Table 1) and compound 2 (Table 2), which were previously characterized as FPR2 agonists in our laboratories, as starting points for the development of FPR2 antagonists with ureidopropanamide structures [28, 33].
Table 1.
Biological Activity of Ureidopropanamides Derived from Modification of Compounds (R)- and (S)-4.
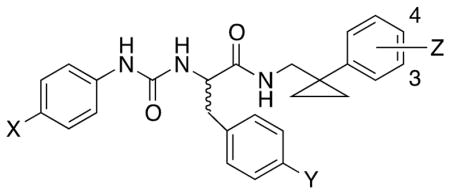
| ||||||||
|---|---|---|---|---|---|---|---|---|
| Ca+2 mobilization | Ca+2 mobilization | MTT | ||||||
| FPR2-HL60 | FPR1-HL60 | FPR2-HL60 | FPR1-HL60 | |||||
| Compd. | X | Y | Z | EC50, μM (efficacy,%)[a] | EC50, μM (efficacy, %)[a] | IC50, μM (max. inhibition, %)[a] | IC50, μM (max. inhibition, %)[a] | EC50, μM[a] |
| (R)-4 | OCH3 | CN | 4-Cl-3-F | 0.63 ± 0.2 (100)[b] | 1.8 ± 0.3 (60) [b] | N.T.[c] | N.T. | N.T. |
| (S)-4 | OCH3 | CN | 4-Cl-3-F | 0.45 ± 0.1 (115) [b] | 3.4 ± 0.9 (60) [b] | N.T. | N.T. | N.T. |
| (R)-24 | OH | CN | H | N.A. [d] | N.A. | N.A. | N.A. | >100 |
| (S)-24 | OH | CN | H | N.A. | N.A. | N.A. | 16.4 ± 3.4 (55) | >100 |
| (R)-25 | OCH3 | OH | H | 0.41 ± 0.16 (90) | 5.1 ± 1.3 (55) | 24.8 ± 2.4 (75) | N.A. | >100 |
| (S)-25 | OCH3 | OH | H | 2.9 ± 0.8 (130) | 1.5 ± 0.3 (95) | 9.8 ± 2.1 (95) | N.A. | >100 |
| (R)-26 | OCH3 | CN | 3-OH | 2.6 ± 0.14 (65) | N.A. | 2.1 ± 0.4 (60) | N.A. | 44.4 |
| (S)-26 | OCH3 | CN | 3-OH | 0.18 ± 0.09 (115) | 1.4 ± 0.4 (100) | 0.98 ± 0.13 (85) | N.A. | >100 |
| (R)-27 | OCH3 | CN | 4-OH | 7.0 ± 2.0 (70) | N.A. | 29.5 ± 2.6 (50) | N.A. | >100 |
| (S)-27 | OCH3 | CN | 4-OH | 1.6 ± 0.5 (75) | 1.3 ± 0.4 (120) | 3.7 ± 1.3 (85) | N.A. | 21.8 |
data are the mean of three indipendent experiments; N.T.= not tested; N.A.= not active;
data taken from ref. [33];
Table 2.
Biological Activity of Ureidopropanamides Derived from Modification of Compound 2.
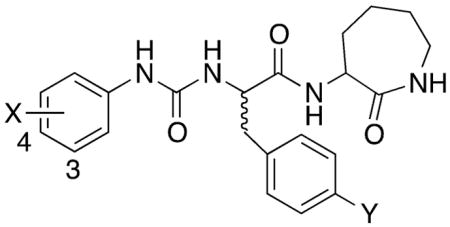
| |||||||
|---|---|---|---|---|---|---|---|
| Ca+2 mobilization | Ca+2 mobilization | MTT | |||||
| FPR2-HL60 | FPR1-HL60 | FPR2-HL60 | FPR1-HL60 | ||||
| Compd. | X | Y | EC50, μM (efficacy, %)[a] | EC50, μM (efficacy, %)[a] | IC50, μM (max. inhibition, %)[a] | IC50, μM (max. inhibition, %)[a] | EC50, μM[a] |
| (S)-2 | 4-Br | H | 0.004 ± 0.002 (115)[b] | 0.3 ± 0.08 (135) [b] | N.T.b | N.T. | N.T. |
| (2R)-42 | 4-OH | H | N.A.c | N.A. | N.A. | N.A. | >100 |
| (2R)-43 | 3-OH | H | N.A. | 12.5 ± 2.5 (90) | N.A. | N.A. | >100 |
| (2S)-44 | 4-OH | H | N.A. | 11.0 ± 3.6 (65) | N.A. | N.A. | >100 |
| (2S)-45 | 3-OH | H | N.A. | 0.55 ± 0.13 (90) | N.A. | 1.2 ± 0.4 (90) | >100 |
| (2R)-46 | 4-Br | OH | 0.35 ± 0.12 (140) | 0.82 ± 0.37 (110) | 25.3 ± 7.2 (100) | 0.56 ± 0.17 (85) | >100 |
| (2S)-47 | 4-Br | OH | 0.78 ± 0.23 (90) | 0.23 ± 0.05 (125) | 2.1 ± 0.6 (100) | 0.076 ± 0.014 (95) | >100 |
data are the mean of three indipendent experiments; N.T.= not tested; N.A.= not active;
data taken from ref. [28].
Results and Discussion
Study Design
The aim of this study was to identify new FPR2 antagonists by structural manipulation of the ureidopropanamide scaffold of the agonists 2 and 4. Many examples have been reported in which small structural modifications to GPCR-targeted ligands lead to major changes in their functional activity, converting agonists into antagonists or vice versa [38]. An example is the FPR2 antagonist Quin C7, in which the replacement of the methoxy group on the 2-phenyl ring of the quinazolinone backbone in Quin C1 with the hydroxyl substituent resulted in the reversal of bioactivity [39]. Therefore, we modified the selected ureidopropanamide agonists 2 and 4 by introducing a hydroxyl group on each aromatic ring (compounds (R)- and (S)-24-27, Table 1 and compounds (2R)-42,43,46 and (2S)-44,45,47 Table 2).
Chemistry
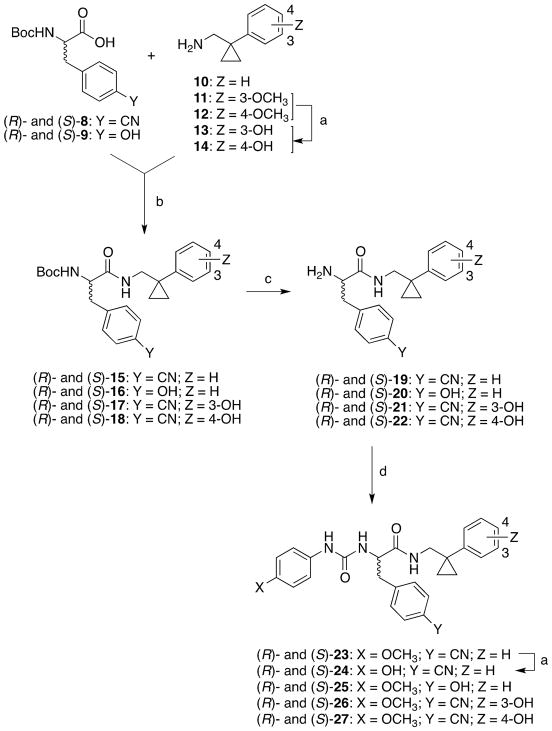
Synthesis of the target compounds (R)- and (S)-24-27 is shown in Scheme 1. Amine 10 was prepared according to reported protocols [39]. Amines 13 and 14 were prepared by demethylation of intermediates 11 and 12 [39], respectively, using BBr3/DCM. Amines 10, 13, and 14 were then condensed with the appropriate (R)-Boc- or (S)-Boc-amino acids 8 and 9 after activation with N-N′-carbonyldiimidazole to obtain the Boc-protected derivatives (R)- and (S)-15-18. Subsequently, these latter compounds were deprotected with 3N hydrochloric acid to obtain amines (R)- and (S)-19-22. The target compounds (R)- and (S)-25-27 were obtained by condensing amines (R)- and (S)-20-21 with the appropriate phenylisocyanate. These compounds were obtained in low yields because of the low solubility of the reagents in the commonly used solvents. Demethylation of compounds (R)- and (S)-23, which were prepared by condensing amines (R)- and (S)-19 with 4-methoxyphenylisocyanate, gave the target compounds (R)- and (S)-24. Also in this case, the reaction proceeded with very low yield because of the low solubility of compounds (R)- and (S)-23 in anhydrous CH2Cl2. The use of different solvents did not lead to a substantial increase of the yield.
Scheme 1.
Synthesis of the Target Compounds (R)- and (S)-24-27.
Reagents and conditions: a) BBr3, DCM, 0 °C to RT, 4 h; b) N,N′-carbonyldiimidazole, RT, overnight; c) 3N HCl, dioxane, RT, 24 h; d) 4-methoxyphenylisocyanate, RT, overnight.
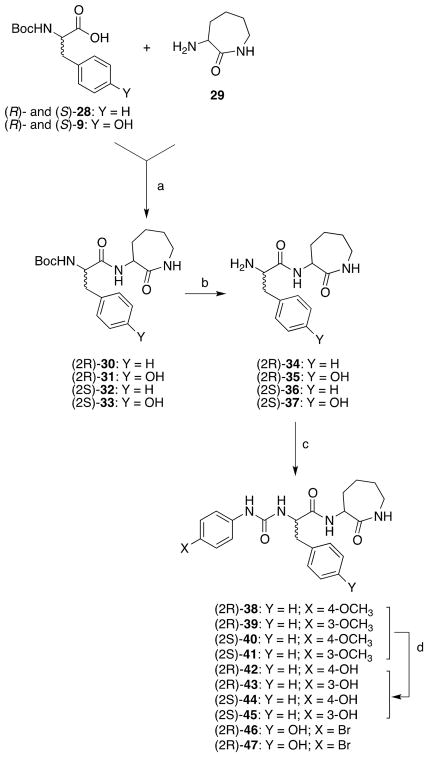
The target compounds (2R)-42,43,46 and (2S)-44,45,47, which are structurally related to compound 2, were prepared in a similar fashion according to Scheme 2. (R)-Boc- or (S)-Boc-phenylalanine (28) or tyrosine (9) was condensed with the commercially available 2-amino-ε-caprolactam (29) after activation with N-N′-carbonyldiimidazole to obtain the Boc-protected derivatives (2R)-30,31 and (2S)-32,33. Subsequently, these latter compounds were deprotected with 3N hydrochloric acid to obtain amines (2R)-34,35 and (2S)-36,37. The target compounds (2R)-46 and (2S)-47 were obtained by condensing amines (2R)-35 and (2S)-37 with 4-bromophenylisocyanate, respectively. For both compounds low yields were observed because of the low solubility of the reagents in the commonly used solvents, such THF or dioxane. Target compounds (2R)-42,43 and (2S)-44,45 were prepared by demethylation of compounds (2R)-38,39 and (2S)-40,41, which were prepared by condensing the amines (2R)-34 and (S)-36 with 4-methoxy or 3-methoxyphenylisocynate, respectively.
Scheme 2.
Synthesis of the Target Compounds (2R)-42,43,46 and (2S)-44,45,47.
Reagents and conditions: a) N,N′-carbonyldiimidazole, RT, overnight; b) 3N HCl, dioxane, RT, 24 h; c) 4-substituted phenylisocyanate, RT, overnight; d) BBr3, DCM, 0 °C to RT, 4 h.
Intrinsic Activity of the Target Compounds
Functional activity of the newly synthetized compounds was assessed in HL-60 cells transfected with human FPR2 by evaluating their effect on intracellular Ca2+ flux. The functional activity at FPR1 was also assessed in HL-60 cells transfected with human FPR1 to determine compound selectivity.
The first group of compounds originated from the agonist (R)- and (S)-4 (Table 1). The data indicate that none of the compounds behaved as FPR2 antagonists, suggesting that the simple introduction of a hydroxyl substituent on one of the aromatic rings of the ureidopropanamide derivatives was not able to convert the functional response from agonism to antagonism. However, introduction of the hydroxyl group did have different impacts on biological activity depending on its location. For example, when the hydroxyl group was introduced in the 4-position of the phenylureidic moiety ((R)- and (S)-24), the compounds did not induce Ca2+ mobilization in FPR2-HL60 cells and did not block Ca2+ mobilization induced by an agonist in these cells, indicating that the compounds were no longer able to interact with FPR2.
This result was quite unexpected because a previous structure-activity study indicated that different substituents, including polar ones such as NO2 or CN, were well tolerated in that position [33]. Thus, it is likely that the introduction of an H-bond donor substituent is not tolerated in this part of the molecule. On the other hand, (R)- and (S)-24 behaved as FPR1 antagonist and inhibited Ca2+ mobilization induced by fMLF.
Replacement of the cyano group of compounds (R)- and (S)-4 with the hydroxyl group was well tolerated. In fact, (R)- and (S)-25 exhibited EC50 values comparable to those of compound (R)- and (S)-4 at both FPR2 and FPR1, confirming our previous findings that a polar group is well tolerated in that position [30, 33]. When the hydroxyl substituent was introduced on the phenyl ring of the “right hand” of the molecule, different effects were observed depending on the position and the chirality of the molecule. Indeed, (R)-enantiomers were less active than their (S)-counterparts. In addition, when the hydroxyl group was introduced at the 3-position, the resulting compound ((S)-26) had an EC50 value comparable to that of compound (S)-4, whereas the 4-substituted derivative ((S)-27) had slightly lower agonist potency as compared to (S)-4. This trend was in line with our previous finding suggesting that the size and position of the substituent in this part of the molecule influences the interaction with FPR2 [33]. Finally, the (R)-enantiomers 26 and 27 were not able to activate FPR1, whereas the (S)-enantiomers had EC50 values comparable to that of compound (S)-4. Analysis of the derivatives of agonist 2 (Table 2) showed that introduction of the hydroxyl substituent on the phenyl linked to the ureidic group led to a complete loss of FPR2 agonist activity and a substantial decrease in agonist potency for FPR1. This effect was more pronounced when the hydroxyl group was placed in 4-position, confirming that the introduction of an H-bond donor substituent in this part of the molecule did not allow a favorable interaction with FPR2. Replacement of the phenylalanine residue in 2 with the tyrosine led to a decrease of FPR2 agonist potency (compounds (2R)-46 and (2S)-47), although the compounds showed EC50 values in the low micromolar range. On the other hand, this structural modification did not influence the interaction with FPR1.
Since none of the compounds behaved as an antagonist, we evaluated if they were able to induce receptor desensitization and, thus, if they could behave functionally as FPR2 antagonists. It is known that after stimulation with N-formyl peptides FPRs undergo homologous desensitization and, as a result, the cellular responses rapidly decline in intensity and the cells become refractory to subsequent stimulation with agonists [9]. Desensitization is one of the mechanisms for controlling and regulating GPCR signaling and trafficking [40], and it has been proposed that targeting desensitization machinery would result in fine tuning of physiological responses [41]. For example, recent studies have demonstrated that LXA4 stimulation induced FPR2 internalization, which is critical for phagocytosis of apoptotic cells [42]. In addition, we previously reported that compound 2 was able to induce receptor desensitization in human neutrophils [28]. Therefore, we evaluated if our compounds were able to induce FPR2 desensitization by inhibiting Ca2+ mobilization induced by the selective FPR2 agonist WKYMVM. Among the compounds generated from compound (R)- and (S)-4, only compounds (R)- and (S)-26-27, in which the hydroxyl group was inserted in the phenyl ring of the “right hand” of the molecule, were able to induce FPR2 desensitization with IC50 values comparable to those of their EC50 values. Regarding compounds (2R)-42,43, 46 and (2S)-44,45,47, formally derived from compound 2, only compounds (2R)-46 and (2S)-47 were able to induce desensitization. Compounds (2S)-45, (2R)-46, and (2S)-47 were able to induce FPR1 desensitization. Again, their IC50 values are comparable to their EC50 values, with the exception of compound (2R)-46 which was less effective at desensitizing FPR2. These data suggest that the agonist-selective desensitization is related to subtle changes in the molecular structure.
Effect of Selected Compounds on ROS Production in N9 Cells
Considering the ability of our FPR2 agonists to induce receptor desensitization, we considered whether they could exert a protective effect in an in vitro model of neuroinflammation. To address this issue, we evaluated their effects in mouse microglia N9 cells, which have been extensively used as a representative model of primary microglial cells [43]. Moreover, it has been reported that N9 cells express low levels of FPR2 mRNA under resting conditions, whereas FPR2 mRNA expression is induced in a time-dependent manner upon cell activation by bacterial endotoxin lipopolysaccharide (LPS) or others inflammatory stimulus, such as β-amyloid [44,45].
Initially, we evaluated the effect of newly synthesized compounds on metabolic activity in N9 cells under resting conditions using the MTT (3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazoliumbromide) assay, which quantifies mitochondrial activity in living cells, in order to assess cytotoxicity of the compounds. The data indicate that none of the compounds, except (R)-26 and (S)-27, were cytotoxic after 24 h treatment in a concentration range of 0.1 to 100 μM (EC50 > 100 μM) (Tables 1 and 2).
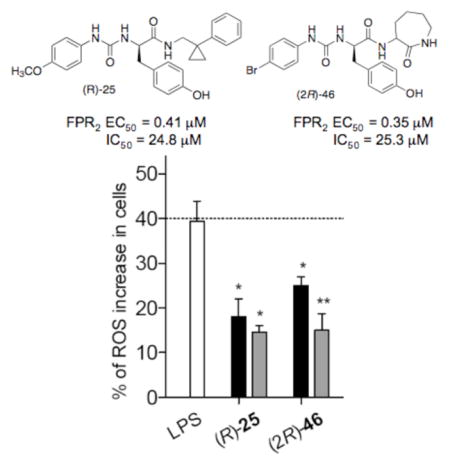
Next, among the studied compounds, we selected the desensitizing compounds (R)-25, (R)- and (S)-26-27 and (2R)-46 and (2S)-47 to evaluate if they were able to modulate oxidant stress in N9 cells induced by an inflammatory stimulus, i.e.: 24 h treatment with LPS. In particular, we measured the effects on ROS production and, for comparative purposes, we included also the reference agonist Quin C1, which is also able to induce FPR2 desensitization (IC50 = 0.04 μM) and displays anti-inflammatory properties in vivo [29]. Under physiological conditions, ROS are involved in immune responses and inflammation, as well as synaptic plasticity, learning and memory [46, 47]. Recently, it has been reported that FPR2 promotes neural differentiation in mouse neural stem cells through ROS generation [48]. However, when produced in excess, ROS can induce oxidative stress, damage proteins and DNA, and induce lipid peroxidation. Oxidative stress can trigger cell death and has been implicated in the pathogenesis of many neurodegenerative diseases, including AD [49].
Assessment of the effect of (R)-25, (R)- and (S)-26-27, (2R)-46 and (2S)-47 and Quin C1 in N9 cells after 24 h stimulation with LPS showed that at a 0.1 μM concentration, only compounds Quin-C1, (R)-25, and (2R)-46 induced a statistically significant reduction in ROS production (p< 0.05) (Figure 2). When the compounds were tested at a 1 μM concentration, a statistically significant reduction of ROS production was observed for all of the tested compounds, except compound (S)-26 (p< 0.05) (Figure 2), indicating dose-dependence of this response. Moreover, except for compound (S)-26, the reduction of ROS production correlated with the levels of receptor desensitization measured for the respective compounds.
Figure 2.
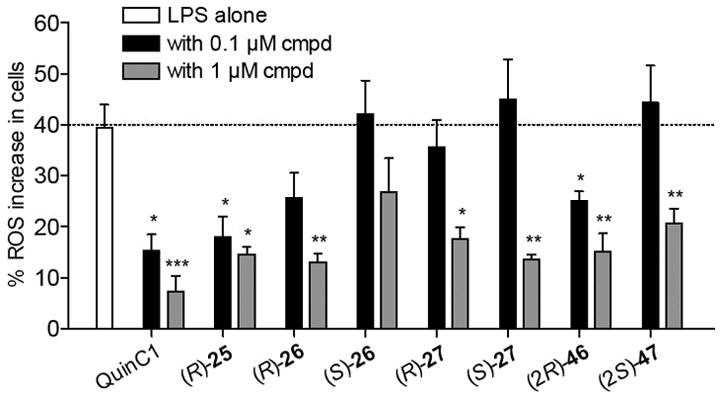
Effect of compounds of selected FPR2 ligands on ROS production in mouse N9 cells. The white bar is the percentage of ROS increase observed after 24 h treatment with LPS as compared to control. The black and grey bars are the percentage of ROS increase observed after 24 h co-incubation of test compound and LPS at 0.1 μM and 1 μM, respectively, as compared to control (p < 0.05).
We also evaluated direct ROS scavenging activity of (R)-25, (R)- and (S)-26-27, (2R)-46 and (2S)-47 using a non-enzymatic O2·−-generating system (phenazine methosulfate/NADH). Importantly, none of the tested compounds demonstrated ROS scavenging activity at concentrations up to 20 μM (data not shown), indicating that (R)- and (S)-26-27, (2R)-46 and (2S)-47 were behaving as functional antagonists and not as ROS scavengers and that they are able to exert protective effects in our model of neuroinflammation through receptor desensitization and possibly cross-desensitization.
Conclusions
In summary, we have manipulated the structure of the FPR2 agonists (R)- and (S)-4 and 2 in order to develop FPR2 antagonists with a ureidopropanamide scaffold. We inserted an hydroxyl group on each aromatic ring individually in an effort to interconvert functional activity from agonism to antagonism, as the same structural modification on the quinazolinone agonist Quin C1 led to the antagonist Quin C7. We prepared and tested fourteen new structurally related ureidopropanamide derivatives. Although none of the compounds behaved as an antagonist, some of the compounds were able to induce receptor desensitization, thus behaving as functional antagonists. Further analysis of compounds (R)-25, (R)- and (S)-26-27, (2R)-46, and (2S)-47 in an in vitro model of neuroinflammation showed that these compounds did not induce inflammatory response in unstimulated mouse microglial N9 cells but were able to significantly reduce the production of ROS when the cells were stimulated with LPS, thereby exerting protective effects against oxidative stress. In particular, derivatives (R)-25, and (2R)-46 induced a dose-dependent reduction in cellular ROS level. These data are very promising because oxidative stress is implicated in the pathogenesis of many neurodegenerative diseases and open new questions if other FPR2 agonists capable to desensitize the receptor can exert protective effect against inflammation-associated oxidative stress. Future studies will assess if these compounds can also exert anti-inflammatory effects by reducing the intracellular levels of pro-inflammatory mediators through the desensitization of FPR2.
Experimental Section
Chemistry
Chemicals were purchased from Sigma-Aldrich, Alfa Aesar, and TCI Chemicals. Unless otherwise stated, all chemicals were used without further purification. Thin layer chromatography (TLC) was performed using plates from Merck (silica gel 60 F254). Column chromatography was performed with 1:30 Merck silica gel 60 Å (63–200 μm) as the stationary phase. Flash chromatographic separations were performed on a Biotage SP1 purification system using flash cartridges pre-packed with KP-Sil 32–63 μm, 60 Å silica gel. 1H NMR spectra were recorded on a Varian Mercury-VX spectrometer (300 MHz) or an Agilent NMR spectrometer (500 MHz). All chemical shift values are reported in ppm (δ). For enantiomeric pairs, NMR spectra of both enantiomers were recorded; however, only the NMR spectrum of the (R)-enantiomer is reported in the experimental section. Recording of mass spectra was performed on an HP6890-5973 MSD gas chromatograph/mass spectrometer; only significant m/z peaks, with their percentage of relative intensity in parentheses are reported. HRMS-ESI analyses were performed on a Bruker Daltonics MicrOTOF-Q II mass spectrometer, mass range 50–800 m/z, electrospray ion source in positive or negative ion mode. All spectra were in accordance with the assigned structures. The purity of the target compounds listed in Table 1 and Table 2 was assessed by RP-HPLC and combustion analysis. All compounds showed ≥ 98% purity. RP-HPLC analysis was performed on an Agilent 1260 Infinity Binary LC System equipped with a diode array detector using a Phenomenex Gemini C-18 column (250 × 4.6 mm, 5 μm particle size). All target compounds were eluted with CH3OH/H2O, 8:2 (v/v) at a flow rate of 1 mL/min. Elemental analyses (C,H,N) of the target compounds were performed on an Eurovector Euro EA 3000 analyzer. Analyses indicated by the symbols of the elements were within ± 0.4 % of the theoretical values. Enantiomeric purity of the target compounds (R)- and (S)-24-27 was assessed by chiral HPLC analysis on a Perkin-Elmer series 200 LC instrument using a Daicel ChiralCel OD column (250 mm × 4.6 mm, 5 μm particle size) and equipped with a Perkin-Elmer 785A UV/VIS detector setting λ = 230 nm. The compounds were eluted with n-hexane/EtOH, 4:1, v/v at a flow rate of 0.8 mL/min. All compounds showed enantiomeric excesses ≥ 98%.
The following compounds were prepared as previously described: [1-(phenyl)cyclopropyl]methanamine (10) [39]; [1-(3-methoxyphenyl)cyclopropyl]methanamine (11) [39]; [1-(4-methoxyphenyl)cyclopropyl]methanamine (12) [39]; (R)- and (S)-tert-butyl {3-(4-cyanophenyl)-1-[[[1-(phenyl)cyclopropyl]methyl]amino]-1-oxopropan-2-yl}carbamate ((R)- and (S)-15) [33]; (R)- and (S)-2-amino-3-(4-cyanophenyl)-N-[(1-phenylcyclopropyl)methyl]propanamide ((R)- and (S)-19) [33].
General Procedure for the Synthesis of Boc-protected Derivatives (R)- and (S)-16 and (2R)-31
N,N′-Carbonyldiimidazole (1.1 mmol) was added to a solution of (R)- or (S)-Boc-protected amino acid (1.0 mmol), in anhydrous THF (10 mL) under N2. The reaction mixture was stirred at room temperature overnight, then a solution of the appropriate amine (1.0 mmol) in anhydrous THF was added. The reaction mixture was stirred at room temperature for 6 h. After removal of the solvent in vacuo, the residue was partitioned between EtOAc (20 mL) and H2O (2 × 20 mL). The aqueous layer was separated and extracted twice with EtOAc (20 mL). The collected organic layers were dried (Na2SO4) and evaporated in vacuo. The crude residue was chromatographed as detailed below to give pure target compound as a white solid.
(R)-tert-Butyl {3-(4-hydroxyphenyl)-1-oxo-1-[[(1-phenylcyclopropyl)methyl]amino]propan-2-yl}carbamate ((R)-16)
Eluted with CHCl3/AcOEt 8:2. 40% Yield. 1H NMR (500 MHz, CD3OD): δ= 0.76–0.88 (m, 4H), 1.37 (s, 9H), 2.63 (dd, 1H, J= 13.7 and 8.3 Hz), 2.75 (dd, 1H, J= 13.7 and 8.3 Hz), 2.85 (dd, 1H, J= 13.7 and 5.9 Hz), 2.95 (dd, 1H, J= 13.7 and 5.9 Hz), 4.13–4.17 (m, 1H), 6.68 (d, 2H, J= 8.3 Hz), 6.74 (br d, 1H), 6.96–6.99 (m, 2H), 7.18 (br t, 1H), 7.23–7.30 ppm (m, 5H). ESI+/MS m/z 433 (M+Na)+, ESI+/MS/MS m/z 333 (100).
(S)-tert-Butyl{3-(4-Hydroxyphenyl)-1-oxo-1-[[(1-phenylcyclopropyl)methyl]amino]propan-2-yl}carbamate ((S)-16)
Eluted with CHCl3/AcOEt 8:2. 30% Yield. ESI+/MS m/z 433 (M+Na)+, ESI+/MS/MS m/z 333 (100).
(2R)-tert-Butyl {3-(4-hydroxyphenyl)-1-oxo-1-[(2-oxoazepan-3-yl)amino]propan-2-yl}carbamate ((2R)-31)
Eluted with CHCl3/MeOH 19:1. 60% Yield. 1H NMR (500 MHz, CDCl3): δ= 1.23–1.49 (m + s, 11H), 1.76–1.81 (m, 3H, 1H D2O exchanged), 1.89–2.07 (m, 2H), 2.92–3.00 (m, 2H), 3.22–3.26 (m, 2H), 4.36–4.46 (m, 2H), 5.14–5.30 (m, 1H), 6.39–6.47 (m, 1H), 6.72–6.74 (m, 2H), 6.85–7.05 (m, 2H), 7.12 ppm (br s, 1H). ESI+/MS m/z 414 (M+Na)+, ESI+/MS/MS m/z 314 (100).
General Procedure for the Synthesis of Amines (R)- and (S)-20 and (2S)-35
To a solution of Boc-protected derivatives (R)- and (S)-16 or (2R)-31 (0.46 mmol) in 1,4-dioxane (10 mL) 3N hydrochloric acid (5 mL) was added. The reaction mixture was stirred at room temperature for 24 h and basified with 5% aqueous NaOH. The separated aqueous phase was extracted with EtOAc (2 × 20 mL). The combined organic layers were dried (Na2SO4) and concentrated in vacuo to afford pure target amine (colorless oil) in quantitative yield.
(R)-2-Amino-3-(4-hydroxyphenyl)-N-[(1-phenylcyclopropyl)methyl]propanamide ((R)-20)
1H NMR (500 MHz, CD3OD): δ= 0.77–0.87 (m, 4H), 2.62 (dd, 1H, J= 13.7, 6.8 Hz), 2.80 (dd, 1H, J= 13.7, 6.4 Hz), 3.30–3.35 (m, 2H), 3.40–3.45 (m, 2H), 6.71 (d, 2H, J= 8.3 Hz), 6.97 (d, 2H, J= 8.3 Hz), 7.16 (br t, 1H), 7.15–7.19 (br t, 1H), 7.23–7.30 ppm (m, 5H). ESI+/MS m/z 333 (M+Na)+, ESI+MS/MS m/z 162 (100), 131 (64).
(S)-2-Amino-3-(4-hydroxyphenyl)-N-[(1-phenylcyclopropyl)methyl]propanamide ((S)-20)
ESI+/MS m/z 333 (M+Na)+, ESI+MS/MS m/z 162 (100), 131 (62).
(2R)-2-Amino-3-(4-hydroxyphenyl)-N-(2-oxoazepan-3-yl)propanamide ((2R)-35)
1H NMR (500 MHz, CD3OD): δ 1.22–1.57 (m, 2H), 1.66–2.05 (m, 4H), 1.98–2.02 (m, 2H), 2.87–3.09 (m, 1H), 3.19–3.30 (m, 3H), 3.51–3.77 (m, 1H), 4.47–4.58 (m, 1H), 6.76 (d, 2H, J= 8.3 Hz), 7.02–7.16 ppm (m, 2H). ESI−/MS m/z 314 (M+Na)+, ESI+MS/MS m/z 84 (100), 120 (49).
General Procedure for the Synthesis of the Compounds (R)- 23,25 and (2R)-46
To a solution of the amine (R)- and (S)-19,20 or (2R)-35 (1.0 mmol) in anhydrous THF, a solution of the appropriate 4-substitued phenylisocyanate (1.2 mmol) in the same solvent (10 mL) was added and the reaction mixture was stirred at room temperature overnight. After removing the solvent in vacuo, the residue was dissolved in CHCl3 (20 mL) and washed with H2O (2 × 20 mL). The separated organic layer was dried over Na2SO4 and concentrated under reduced pressure. The crude residue was chromatographed as detailed below. When necessary, the obtained solid was further purified by crystallization from MeOH to give the pure target compound.
(R)-3-(4-Cyanophenyl)-2-[3-(4-methoxyphenyl)ureido]-N-[(1-phenylcyclopropyl)methyl]propanamide ((R)-23)
Eluted with CHCl3/AcOEt 8:2. 47% Yield. 1H NMR (500 MHz, [D6]DMSO): δ= 0.69–0.71 (m, 1H), 0.77–0.79 (m, 1H), 0.82–0.85 (m, 2H), 2.76 (dd, 1H, J= 13.2, 7.8 Hz), 2.92 (dd, 1H, J= 13.7, 4.9 Hz), 3.18 (dd, 1H, J= 13.7, 4.9 Hz), 3.47 (dd, 1H, J= 13.7, 6.4 Hz), 3.67 (s, 3H), 4.51 (dd, 1H, J= 13.2, 7.8 Hz), 6.20 (d, 1H, J= 8.3 Hz), 6.78 (d, 2H, J= 8.8 Hz), 7.14–7.16 (m, 1H), 7.19–7.26 (m, 8H), 7.67 (d, 2H, J= 8.8 Hz), 8.15 (br t, 1H), 8.42 ppm (s, 1H). ESI+/MS m/z 491 (M+Na)+, ESI+/MS/MS m/z 491 (100), 342 (20).
(R)-3-(4-Hydroxyphenyl)-2-[3-(4-methoxyphenyl)ureido]-N-[(1-phenylcyclopropyl)methyl]propanamide ((R)-25)
Eluted with CHCl3/MeOH 19:1. 17% Yield. 1H NMR (500 MHZ, CDCl3): δ= 0.74–0.79 (m, 4H), 1.71 (br s, 1H, D2O exchanged), 2.80–2.87 (m, 2H), 3.24 (dd, 1H, J= 13.7, 4.4 Hz), 3.41 (dd, 1H, J= 13.7, 5.9 Hz), 3.72 (s, 3H), 4.46–4.50 (m, 1H), 5.95 (br s, 1H), 6.51 (br t, 1H), 6.62 (d, 2H, J= 7.8 Hz), 6.73 (d, 2H, J= 8.3 Hz), 6.87 (d, 2H, J= 8.3 Hz), 7.02 (d, 2H, J= 7.8 Hz), 7.12–7.16 (m, 4H), 7.20–7.23 ppm (m, 2H). ESI+/MS m/z 482 (M+Na)+, ESI+/MS/MS m/z 482 (100), 333 (92). Anal. calcd for C27H29N3O4: C 70.57, H 6.36, N 9.14, found: C 70.53, H 6.53, N 9.04
(2R)-2-[3-(4-Bromophenyl)ureido]-3-(4-hydroxyphenyl)-N-(2-oxoazepan-3-yl)propanamide ((2R)-46)
Eluted with CHCl3/MeOH 98:2. 22% Yield. 1H NMR (500 MHz, [D6]DMSO): δ= 1.16–1.20 (m, 1H), 1.27–1.39 (m, 1H), 1.59–1.87 (m, 4H), 2.67–2.74 (m, 1H), 2.81–2.93 (m, 1H), 3.01–3.05 (m, 1H), 3.11–3.20 (m, 1H), 4.33–4.39 (m, 1H), 4.40–4.47 (m, 1H), 6.28–6.36 (m, 1H), 6.61 (d, 2H, J= 8.3 Hz), 7.00 (dd, 2H, J= 8.8, 3.4 Hz), 7.29 (dd, 2H, J= 8.8, 1.5 Hz), 7.34 (d, 2H, J= 8.8 Hz), 7.81 (br s, 1H), 7.92–8.01 (m, 1H), 8.84 (br d, 1H), 9.14 ppm (s, 1H). ESI+/MS m/z 511 (M+Na)+, ESI+/MS/MS m/z 340 (22), 314 (100). Anal. calcd for C22H25BrN4O4: C 54.00, H 5.15, N 11.46, found: C 54.14, H 5.24, N 11.34.
(R)-3-(4-Cyanophenyl)-2-[3-(4-hydroxyphenyl)ureido]-N-[(1-phenylcyclopropyl)methyl]propanamide ((R)-24)
To a cooled solution of the methoxy derivatives (R)-23, (2.0 mmol) in anhydrous CH2Cl2 (10 mL), 1.0 M boron tribromide in CH2Cl2 (3.45 mL, 3.45 mmol) was added dropwise. The reaction mixture was stirred at room temperature for 4 h and basified with 10% aqueous NH4OH. The separated aqueous phase was extracted with CH2Cl2 (2 × 20 mL). The combined organic layers were dried (Na2SO4) and concentrated under reduced pressure. The crude residue was chromatographed using CHCl3/MeOH 98:2 as eluent detailed to give the target compound as solid (5% yield). 1H NMR (500 MHz, [D6]DMSO): δ= 0.69–0.71 (m, 1H), 0.76–0.79 (m, 1H), 0.81–0.84 (m, 2H), 2.76 (dd, 1H, J= 13.2, 7.8 Hz), 2.92 (dd, 1H, J= 13.7, 4.9 Hz), 3.18 (dd, 1H, J= 14.2, 4.9 Hz), 3.47 (dd, 1H, J= 13.7, 6.4 Hz), 4.48–4.52 (m, 1H), 6.14 (d, 1H, J= 8.3 Hz), 6.60 (d, 2H, J= 8.8 Hz), 7.07 (d, 2H, J= 8.3 Hz), 7.11–7.17 (m, 1H), 7.22–7.25 (m, 6H), 7.67 (d, 2H, J= 7.8 Hz), 8.13–8.15 (m, 1H), 8.27 (s, 1H), 8.93 ppm (s, 1H). ESI+/MS m/z 477 (M+Na)+, ESI+/MS/MS m/z 477 (100), 342 (10). Anal. calcd for C27H26N4O3: C 71.35, H 5.77, N 12.33, found: C 71.52, H 5.74, N 12.34.
Biological Methods
Materials
Black 96-wells clear bottom plates were purchased from PerkinElmer Life and Analytical Sciences (Boston, MA, USA). Cell culture reagents were purchased from EuroClone (Milan, Italy). LPS (lipopolysaccharide from Escherichia coli O111:B4), MTT, H2DCF-DA (2′,7′-dichlorofluorescein diacetate), Iscove’s Modified Dulbecco’s Medium were obtained from Sigma-Aldrich (Milan, Italy). Phenazine methosulfate, NADH, and nitro blue tetrazolium (NBT) were purchased from Sigma Chemical Co. (St. Louis, MO).
Cell Culture
Human promyelocytic leukemia HL-60 cells stably transfected with FPR1 (FPR1-HL-60 cells) or FPR2 (FPR2-HL-60 cells) (kind gifts from Dr. Marie-Josephe Rabiet, INSERM, Grenoble, France) were cultured in RPMI 1640 medium supplemented with 10% heat-inactivated fetal calf serum, 10 mM HEPES, 100 μg/mL streptomycin, 100 U/mL penicillin, and G418 (1 mg/mL), as described previously [30]. Wild-type HL-60 cells were cultured under the same conditions, but without G418.
Murine N9 microglia cells were purchased from Neuro-Zone, Bresso, Italy. N9 cells were grown in Iscove’s Modified Dulbecco’s Medium supplemented with 5% fetal bovine serum, 2 mM glutamine, 100 U/mL penicillin, 100 mg/mL streptomycin, in a humidified incubator at 37 °C with a 5 % CO2 atmosphere.
Ca2+ Mobilization Assay
Changes in intracellular Ca2+ in HL-60 cells were measured with a FlexStation 3 scanning fluorometer (Molecular Devices, Sunnyvale, CA, USA), as described previously [30]. All active compounds were evaluated in parent (wild-type) HL-60 cells to verify that the agonists are inactive in non-transfected cells (not shown). HL-60 cells were suspended in HBSS− containing 10 mM HEPES, loaded with Fluo-4 AM dye (Invitrogen, Carlsbad, CA, USA) (1.25 μg/mL final concentration), and incubated for 30 min in the dark at 37 °C. After dye loading, the cells were washed with HBSS− containing 10 mM HEPES, resuspended in HBSS containing 10 mM HEPES and Ca2+ and Mg2+ (HBSS+), and aliquotted into the wells of flat-bottomed, half-area-well black microtiter plates (2 × 105 cells/well). The compounds of interest were automatically added from a source plate containing dilutions of test compounds in HBSS+, and changes in fluorescence were monitored (λex= 485 nm, λem= 538 nm) every 5 s for 240 s at room temperature. In desensitization experiments, the dye-loaded cells were pretreated with vehicle (DMSO) or different concentrations of the test compounds, and Ca2+ mobilization was monitored. The same wells were then treated with fMLF (5 nM) for FPR1-HL-60 cells or WKYMVM (5 nM) for FPR2-HL-60 cells, and Ca2+ mobilization was monitored after this second treatment to evaluate desensitization of the response to control peptides. The maximum change in fluorescence, expressed in arbitrary units over baseline, was used to determine agonist response. Responses were normalized to the response induced by 5 nM fMLF for FPR1-HL-60 cells or 5 nM WKYMVM for FPR2-HL-60 cells, which were assigned a value of 100%. Each compound was tested in triplicate. Curve fitting (5–6 points) and calculation of median effective concentration values (EC50) were performed by nonlinear regression analysis of the dose–response curves generated using Prism 5 (GraphPad Software Inc., San Diego, CA, USA).
Cytotoxicity Assay
Compound cytotoxicity in N9 cells was determined by evaluating effects on cell growth using the MTT assay at 24 h [50, 51]. On day 1, 20,000 cells/well were seeded into 96-well plates in the final volume of 100 μL. On day 2, various concentrations of the test compounds were added (0.1–100 mM). The potential cytotoxic effect of DMSO, which was used to solubilize the test compounds, was also evaluated. After 24 h incubation with the compounds, MTT (0.5 mg/mL) was added to each well. The plates were incubated at 37 °C for 3–4 h, then the supernatant was removed. The formazan crystals were solubilized using 100 mL of DMSO/EtOH (1:1), and the absorbance values at 570 nm and 630 nm were measured with a Victor 3 microplate reader (PerkinElmer Life Sciences). EC50 values were determined by fitting the absorbance increase percentage versus log[concentration]. Each compound was tested in triplicate at 5–6 concentrations (10−8–10−4 M). Nonlinear curve fitting was performed using Prism 6.03 (GraphPad Software Inc., San Diego, CA, USA).
ROS assay
Evaluation of the effect of test compounds on ROS in N9 cells was performed as described by Hornick et al. with minor modifications [52]. 25,000 cells/well were seeded into black 96-well clear bottom plates in 100 μL medium. After 3–4 h, LPS (300 ng/mL), alone or in combination with the tested compounds (0.1 or 1 μM) of test compounds, was added to the cells. The plates were incubated at 37 °C for 24 h. H2DCF-DA was added in 100 μL of medium to yield a final concentration of 10 μM, and the plates were incubated for additional 30 min. The wells were washed 3 times with ice cold PBS, saline was added, and the fluorescence signals were read with a Tecan Infinity M1000 plate reader using excitation and emission wavelengths of 480 nm and 520 nm, respectively. The increase in fluorescence with respect to basal level was measured. Statistical analyses and data plotting were performed using Prism 6.03 (GraphPad Software Inc., San Diego, CA, USA). Results were expressed as mean ± standard error of at least three biological replicates. Differences in ROS production were analyzed using one-way ANOVA to identify differences and confirmed with paired two tailed t-tests. A p-value < 0.05 was considered significant.
Evaluation of ROS scavenging activity in cell-free non-enzymatic system
O2·− was generated using a non-enzymatic system in the presence or absence of test compounds, and O2·− was detected by monitoring reduction of NBT to monoformazan dye at 560 nm, as described previously [53]. The O2·− -generating system contained 3 mM phenazine methosulfate, 200 mM NADH, and 50 mM NBT in 0.05 M phosphate buffer (pH 7.5). The reactions were monitored at 560 nm with a SpectraMax Plus microplate spectrophotometer at 25 °C, and the rate of absorption change was determined.
Supplementary Material
Acknowledgments
This research was supported in part by National Institutes of Health IDeA Program COBRE Grant GM110732; USDA National Institute of Food and Agriculture Hatch project 1009546; Montana University System Research Initiative: 51040-MUSRI2015-03; and the Montana State University Agricultural Experiment Station.
Footnotes
Conflict of Interest
The authors declare that there are no conflicts of interest.
The Supporting Information contains synthethic procedures and spectroscopic data for Boc-protected derivatives (R)- and (S)-17-18, (2R)-30 and (2S)- 32,33; amines (R)- and (S)-21-22, (2R)-34 and (2S)-36,37; compounds (R)- and (S)-23,25-27, (2R)-38,39,46 and (2S)-40,41,47; compounds (S)-24, (2R)-42,43 and (2S)-44,45.
References
- 1.Kulkarni OP, Lichtnekert J, Anders HJ, Mulay SR. Mediators Inflamm. 2016;2016:2856213. doi: 10.1155/2016/2856213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shabab T, Khanabdali R, Moghadamtousi SZ, Kadir HA, Mohan G. Int J Neurosci. 2016;127:1–29. doi: 10.1080/00207454.2016.1212854. [DOI] [PubMed] [Google Scholar]
- 3.Ransohoff RM. Science. 2016;353:777–783. doi: 10.1126/science.aag2590. [DOI] [PubMed] [Google Scholar]
- 4.Chen WW, Zhang X, Huang WJ. Mol Med Rep. 2016;13:3391–3396. doi: 10.3892/mmr.2016.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hong H, Kim BS, Im HI. Int Neurourol J. 2016;20(Suppl 1):S2–7. doi: 10.5213/inj.1632604.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Serhan CN, Brain SD, Buckley CD, Gilroy DW, Haslett C, O’Neill LA, Perretti M, Rossi AG, Wallace JL. FASEB J. 2007;21:325–332. doi: 10.1096/fj.06-7227rev. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Serhan CN, Hong S, Gronert K, Colgan SP, Devchand PR, Mirick G, Moussignac RL. J Exp Med. 2002;196:1025–1037. doi: 10.1084/jem.20020760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serhan CN, Krishnamoorthy S, Recchiuti A, Chiang N. Curr Top Med Chem. 2011;11:629–647. doi: 10.2174/1568026611109060629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ye RD, Boulay F, Wang JM, Dahlgren C, Gerard C, Parmentier M, Serhan CN, Murphy PM. Pharmacol Rev. 2009;61:119–161. doi: 10.1124/pr.109.001578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boulay F, Tardif M, Brouchon L, Vignais P. Biochemistry. 1990;29:11123–11133. doi: 10.1021/bi00502a016. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez V, Coto E, Setien F, Gonzalez-Roces S, Lopez-Larrea C. Immunogenetics. 1996;44:446–452. doi: 10.1007/BF02602806. [DOI] [PubMed] [Google Scholar]
- 12.Gao JL, Murphy PM. J Biol Chem. 1993;268:25395–25401. [PubMed] [Google Scholar]
- 13.Lacy M, Jones J, Whittemore SR, Haviland DL, Wetsel RA, Barnum SR. J Neuroimmunol. 1995;61:71–78. doi: 10.1016/0165-5728(95)00075-d. [DOI] [PubMed] [Google Scholar]
- 14.Cui Y, Le Y, Yazawa H, Gong W, Wang JM. J Leukoc Biol. 2002;72:628–635. [PubMed] [Google Scholar]
- 15.Svensson CI, Zattoni M, Serhan CN. J Exp Med. 2007;204:245–252. doi: 10.1084/jem.20061826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang G, Zhang L, Chen X, Xue X, Guo Q, Liu M, Zhao J. Sci Rep. 2016;6:25946. doi: 10.1038/srep25946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fiore S, Maddox JF, Perez HD, Serhan CN. J Exp Med. 1994;180:253–260. doi: 10.1084/jem.180.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Perretti M, Leroy X, Bland EJ, Montero-Melendez T. Trends Pharmacol Sci. 2015;36:737–755. doi: 10.1016/j.tips.2015.07.007. [DOI] [PubMed] [Google Scholar]
- 19.Romano M, Cianci E, Simiele F, Recchiuti A. Eur J Pharmacol. 2015;760:49–63. doi: 10.1016/j.ejphar.2015.03.083. [DOI] [PubMed] [Google Scholar]
- 20.Martini AC, Berta T, Forner S, Chen G, Bento AF, Ji RR, Rae GA. J Neuroinflammation. 2016;13:75. doi: 10.1186/s12974-016-0540-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo Z, Hu Q, Xu L, Guo ZN, Ou Y, He Y, Yin C, Sun X, Tang J, Zhang JH. Stroke. 2016;47:490–497. doi: 10.1161/STROKEAHA.115.011223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Medeiros R, Kitazawa M, Passos GF, Baglietto-Vargas D, Cheng D, Cribbs DH, LaFerla FM. Am J Pathol. 2013;182:1780–1789. doi: 10.1016/j.ajpath.2013.01.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ries M, Loiola R, Shah UN, Gentleman SM, Solito E, Sastre M. J Neuroinflammation. 2016;13:234. doi: 10.1186/s12974-016-0692-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jones RM, Neish AS. Free Radic Biol Med. 2017;105:41–47. doi: 10.1016/j.freeradbiomed.2016.10.495. [DOI] [PubMed] [Google Scholar]
- 25.Schepetkin IA, Khlebnikov AI, Giovannoni MP, Kirpotina LN, Cilibrizzi A, Quinn MT. Curr Med Chem. 2014;21:1478–504. doi: 10.2174/0929867321666131218095521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Corminboeuf O, Leroy X. J Med Chem. 2015;58:537–559. doi: 10.1021/jm501051x. [DOI] [PubMed] [Google Scholar]
- 27.Burli RW, Xu H, Zou X, Muller K, Golden J, Frohn M, Adlam M, Plant MH, Wong M, McElvain M, Regal K, Viswanadhan VN, Tagari P, Hungate R. Bioorg Med Chem Lett. 2006;16:3713–3718. doi: 10.1016/j.bmcl.2006.04.068. [DOI] [PubMed] [Google Scholar]
- 28.Schepetkin IA, Kirpotina LN, Khlebnikov AI, Jutila MA, Quinn MT. Mol Pharmacol. 2011;79:77–90. doi: 10.1124/mol.110.068288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He M, Cheng N, Gao WW, Zhang M, Zhang YY, Ye RD, Wang MW. Acta Pharmacol Sin. 2011;32:601–610. doi: 10.1038/aps.2011.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schepetkin IA, Kirpotina LN, Khlebnikov AI, Leopoldo M, Lucente E, Lacivita E, De Giorgio P, Quinn MT. Biochem Pharmacol. 2013;85:404–416. doi: 10.1016/j.bcp.2012.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lacivita E, Schepetkin IA, Stama ML, Kirpotina LN, Colabufo NA, Perrone R, Khlebnikov AI, Quinn MT, Leopoldo M. Bioorg Med Chem. 2015;23:3913–3924. doi: 10.1016/j.bmc.2014.12.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lacivita E, Stama ML, Maeda J, Fujinaga M, Hatori A, Zhang MR, Colabufo NA, Perrone R, Higuchi M, Suhara T, Leopoldo M. Chem Biodivers. 2016;13:875–873. doi: 10.1002/cbdv.201500281. [DOI] [PubMed] [Google Scholar]
- 33.Stama ML, Œlusarczyk J, Lacivita E, Kirpotina LN, Schepetkin IA, Chamera K, Riganti C, Perrone R, Quinn MT, Basta-Kaim A, Leopoldo M. J Med Chem. doi: 10.1016/j.ejmech.2017.09.023. Submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skovbakke SL, Winther M, Gabl M, Holdfeldt A, Linden S, Wang JM, Dahlgren C, Franzyk H, Forsman H. Biochem Pharmacol. 2016;119:56–65. doi: 10.1016/j.bcp.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 35.Zhou C, Zhang S, Nanamori M, Zhang Y, Liu Q, Li N, Sun M, Tian J, Ye PP, Cheng N, Ye RD, Wang MW. Mol Pharmacol. 2007;72:976–983. doi: 10.1124/mol.107.037564. [DOI] [PubMed] [Google Scholar]
- 36.Pinilla C, Edwards BS, Appel JR, Yates-Gibbins T, Giulianotti MA, Medina-Franco JL, Young SM, Santos RG, Sklar LA, Houghten RA. Mol Pharmacol. 2013;84:314–324. doi: 10.1124/mol.113.086595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stepniewski TM, Filipek S. Bioorg Med Chem. 2015;23:4072–4081. doi: 10.1016/j.bmc.2015.03.062. [DOI] [PubMed] [Google Scholar]
- 38.Dosa PI, Amin EA. J Med Chem. 2016;59:810–840. doi: 10.1021/acs.jmedchem.5b00982. [DOI] [PubMed] [Google Scholar]
- 39.Wong YC, Ke Z, Yeung YY. Org Lett. 2015;17:4944–4947. doi: 10.1021/acs.orglett.5b02557. [DOI] [PubMed] [Google Scholar]
- 40.Kelly E, Bailey CP, Henderson G. Br J Pharmacol. 2008;153:S379–S388. doi: 10.1038/sj.bjp.0707604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gainetdinov RR, Premont RT, Bohn LM, Lefkowitz RJ, Caron MG. Annu Rev Neurosci. 2004;27:107–144. doi: 10.1146/annurev.neuro.27.070203.144206. [DOI] [PubMed] [Google Scholar]
- 42.Maderna P, Cottell DC, Toivonen T, Dufton N, Dalli J, Perretti M, Godson C. FASEB J. 2010;24:4240–4249. doi: 10.1096/fj.10-159913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ferrari D, Villalba M, Chiozzi P, Falzoni S, Ricciardi-Castagnoli P, Di Virgilio F. J Immunol. 1996;156:1531–1539. [PubMed] [Google Scholar]
- 44.Tiffany HL, Lavigne MC, Cui YH, Wang JM, Leto TL, Gao JL, Murphy PM. J Biol Chem. 2001;276:23645–23652. doi: 10.1074/jbc.M101031200. [DOI] [PubMed] [Google Scholar]
- 45.Cui YH, Le Y, Gong W, Proost P, Van Damme J, Murphy WJ, Wang JM. J Immunol. 2002;168:434–442. doi: 10.4049/jimmunol.168.1.434. [DOI] [PubMed] [Google Scholar]
- 46.Veal EA, Day AM, Morgan BA. Molecular Cell. 2007;26:1–14. doi: 10.1016/j.molcel.2007.03.016. [DOI] [PubMed] [Google Scholar]
- 47.Kishida KT, Klann E. Antioxid Redox Signal. 2007;9:233–244. doi: 10.1089/ars.2007.9.ft-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhang L, Wang G, Chen X, Xue X, Guo Q, Liu M, Zhao J. Sci Rep. 2017;7:206. doi: 10.1038/s41598-017-00314-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leuner K, Schutt T, Kurz C, Eckert SH, Schiller C, Occhipinti A, Mai S, Jendrach M, Eckert GP, Kruse SE, Palmiter RD, Brandt U, Dröse S, Wittig I, Willem M, Haass C, Reichert AS, Müller WE. Antioxid Redox Signal. 2012;16:1421–1433. doi: 10.1089/ars.2011.4173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colabufo NA, Berardi F, Contino M, Niso M, Abate C, Perrone R, Tortorella V. Naunyn-Schmiedeberg’s Arch Pharmacol. 2004;370:106–113. doi: 10.1007/s00210-004-0961-2. [DOI] [PubMed] [Google Scholar]
- 51.Azzariti A, Colabufo NA, Berardi F, Porcelli L, Niso M, Simone MG, Perrone R, Paradiso A. Mol Cancer Ther. 2006;5:1807–1816. doi: 10.1158/1535-7163.MCT-05-0402. [DOI] [PubMed] [Google Scholar]
- 52.Hornick JR, Xu J, Vangveravong S, Tu Z, Mitchem JB, Spitzer D, Goedegebuure P, Mach RH, Hawkins WG. Mol Cancer. 2010;9:298. doi: 10.1186/1476-4598-9-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schepetkin I, Potapov A, Khlebnikov A, Korotkova E, Lukina A, Malovichko G, Kirpotina L, Quinn MT. J Biol Inorg Chem. 2006;11:499–513. doi: 10.1007/s00775-006-0101-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.