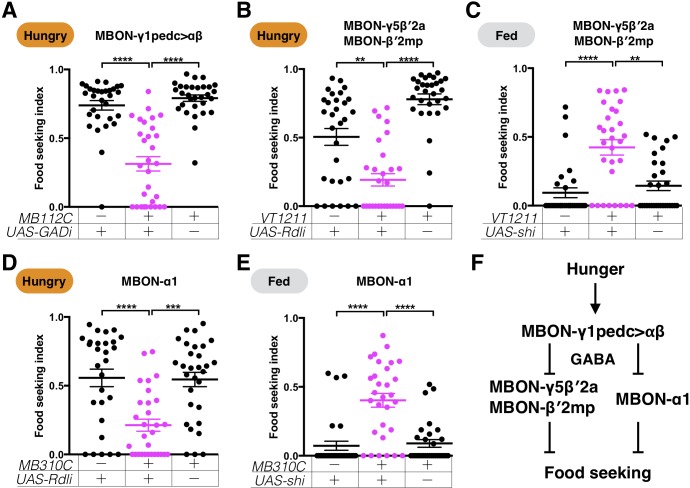
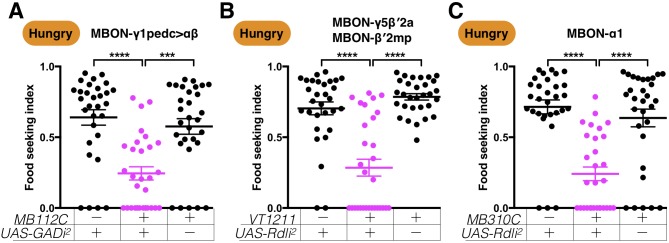
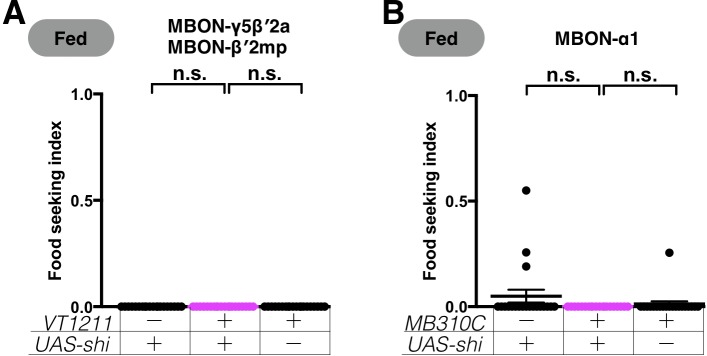
Figure 6. GABAergic MBON-γ1pedc>αβ promotes yeast food-seeking behavior by inhibiting β′2-innervating MBONs and MBON-α1.
Male flies starved for 24 hr (A, B and D) or food-satiated (C and E) were assessed for their yeast food-seeking performance. Individual data points and mean ± SEM are shown. (A) The performance of MB112C;UAS-GAD-RNAi flies was statistically lower than the controls (Kruskal-Wallis, n = 30, p<0.0001). (B) The performance of VT1211-GAL4;UAS-Rdl-RNAi flies was significantly lower than the controls (Kruskal-Wallis, n = 30, p=0.0053). (C) The performance of VT1211-GAL4;UAS-shits1 flies was statistically higher than the controls at a restrictive 32°C (Kruskal-Wallis, n = 30, p=0.0015). (D) The performance of MB310C;UAS-Rdl-RNAi flies was statistically lower than the controls (Kruskal-Wallis, n = 28–30, p=0.0004). (E) The performance of MB310C;UAS-shits1 flies was higher than the controls at a restrictive 32°C (Kruskal-Wallis, n = 30, p<0.0021). (F) A model showing the relationship between MBON-γ1pedc>αβ, MBON-γ5β′2a, MBON-β′2mp, and MBON-α1.