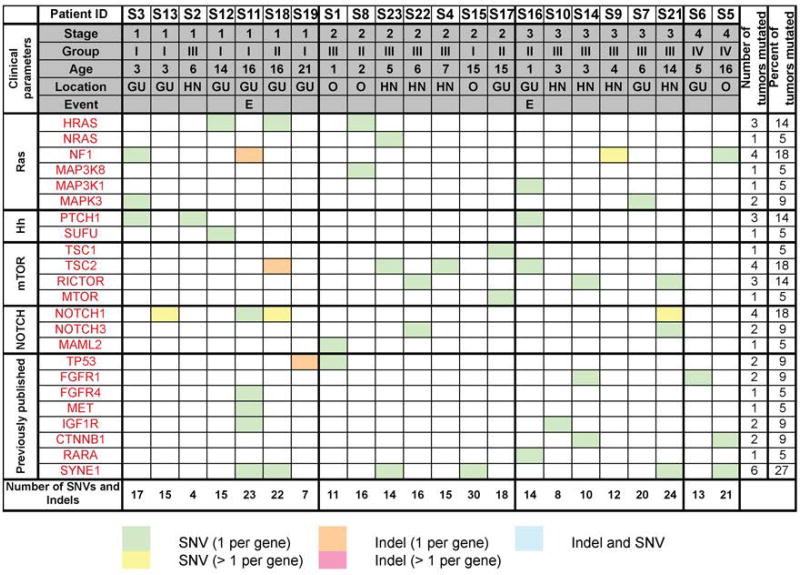
Figure 4. Mutational spectrum of PF- D3 RMS.
Mutations in RAS, NOTCH, Hh and mTOR pathway genes and in genes previously found mutated in RMS3, 4. SNVs were considered if they were non-synonymous, detected in a tumor fraction >30%, not present in normal muscle samples and listed in the Exome Variant Server, NHLBI Exome Sequencing Project (ESP), Seattle, WA, USA, with a population frequency < 1%). InDels were considered if allele frequency was > 25%. The number of SNVs and Indels refers to the total number of aberrations observed in the tumor. Of note, only 2 of 3 recurrent cases were included in the sequencing analysis; one recurrent case was excluded because of insufficient DNA availability.