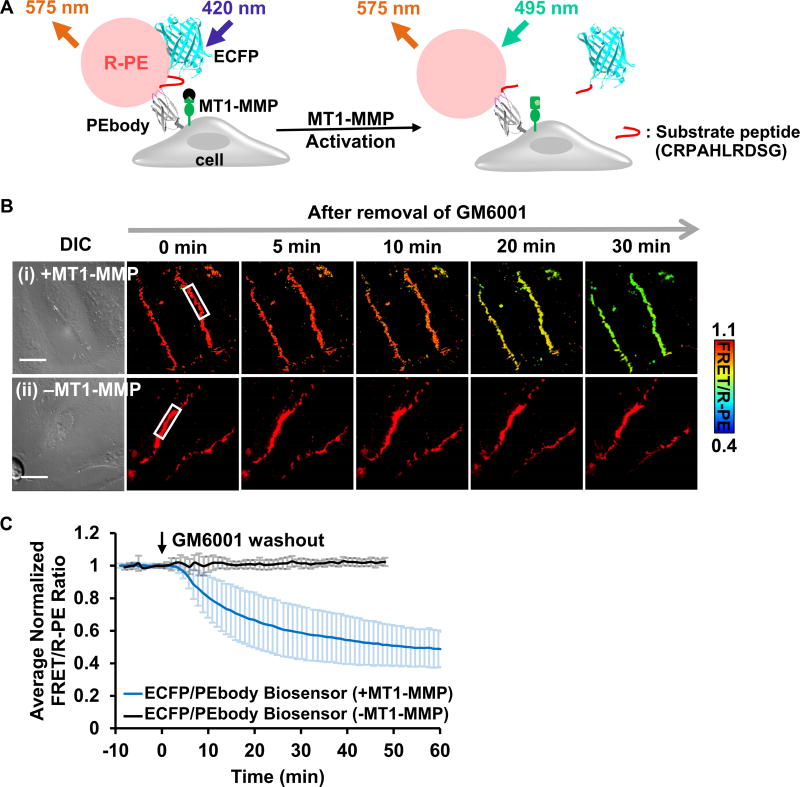
Figure 4. The ECFP/PEbody hybrid FRET biosensor for visualizing MT1-MMP activity on the surface of live cancer cells.
(A) The design strategy and the sensing mechanism of the biosensor. The biosensor was anchored on the extracellular surface using the transmembrane domain of PDGFR. The N-terminus of the transmembrane domain was linked to PEbody to allow for the R-PE binding. A MT1-MMP substrate sequence flanked by flexible linkers was inserted between ECFP and PEbody. Left: R-PE-staining of the intact biosensor allowed the energy transfer from ECFP to R-PE; Right: Following activation, MT1-MMP cleaved the biosensor substrate sequence, disrupted FRET, and reduced the FRET/R-PE ratio. (B) The DIC and FRET/R-PE ratio images before and after GM6001 inhibitor washout of the representative HeLa cells expressing the MT1-MMP ECFP/PEbody biosensor (i) with or (ii) without the full-length MT1-MMP gene. The color bar indicates the FRET/R-PE ratio, with hot and cold colors representing the high and low ratios, respectively. The white boxes indicate the region where the ratio was quantified for (C). Scale bar: 20 µm. (C) The average quantified time course of the normalized FRET/R-PE ratio from multiple HeLa cells in the GM6001 washout assay was compared in cells with and without the full-length MT1-MMP gene. Data are normalized with an individual basal FRET/R-PE ratio before GM6001 washout and represented as mean ± SD (n = 64 and 29; n is the number of analyzed cell contacts with and without the full-length MT1-MMP, respectively). See also Figure S3.