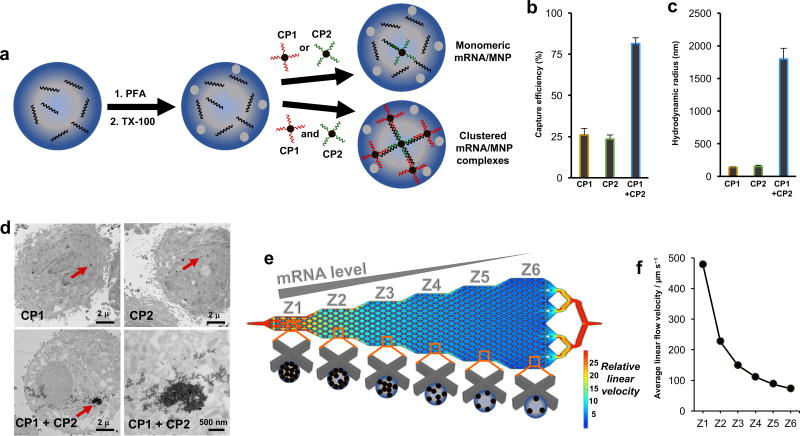
Figure 1. Cellular mRNA analysis approach.
(A) Cells are fixed with 4% paraformaldehyde (PFA) and permeabilized with 0.3% Triton X-100 (TX-100). The cells are incubated with two capture probes (CPs), which are composed of magnetic nanoparticles (MNPs) conjugated to DNA sequences complementary to the target mRNA. Clusters of MNPs are formed and trapped within the cells if two independent CPs are used. (B) When PC3 cells are subjected to magnetic capture based on the targeting of survivin mRNA, only low levels of cell capture are observed if single capture probes are used, while when two capture probes are coincubated with the cells, capture efficiency is increased significantly. (C) DLS measurements of the hydrodynamic radius of magnetic nanoparticles subsequent to hybridization of survivin RNA with either individual capture probes or the combined probes. Statistical analyses of data are provided in Supplementary tables S2−S7. (D) TEM images of PC3 cells after targeting survivin mRNA with CP1, CP2, or CP1+CP2. (E) Device featuring six sequential zones that feature different average linear flow velocities (1×, 0.47×, 0.31×, 0.23×. 0.18×, 0.15×) to facilitate capturing cells with different magnetic content. Cells with high magnetic content are captured in the first zone, whereas cells with medium to low magnetic content are captured in later zones. (F) Distribution of linear velocities at a flow rate of 600 µL h−1 for Zone 1 – 6.