Fig. 5.
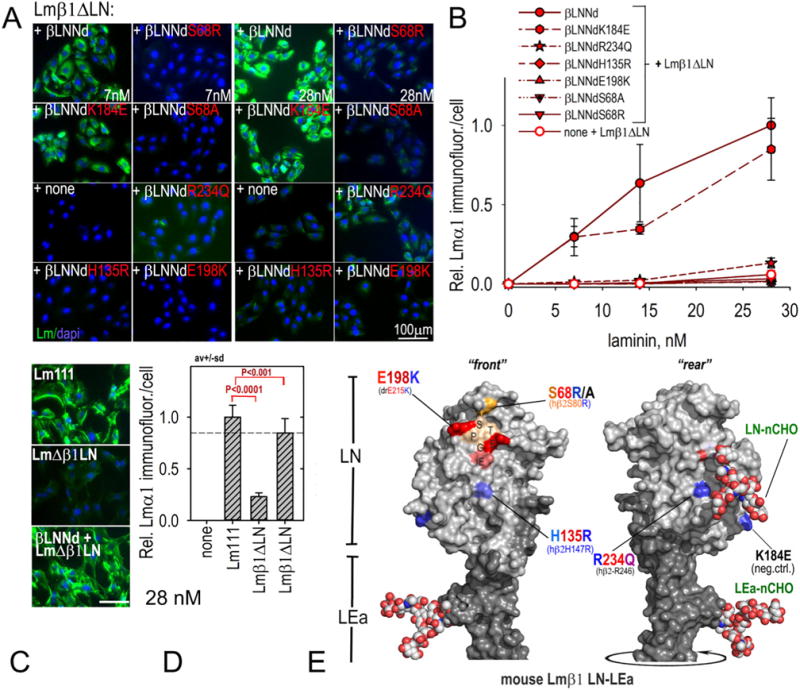
Assembly of βLNNd–Lmβ1ΔLN complexes on cell surfaces. Panels A–D. βLNNd proteins, WT and bearing LN point mutations and coupled to Lmβ1ΔLN were added to the medium of SCs for 1 h, washed, fixed and immunostained to detect cell-adherent laminin through detection of laminin α1. Representative images (panel A) and plots of average (±S.D., n = 6–7 fields/condition) intensity/cell as a function of laminin/βLNNd concentration (panel B) are shown. WT Lm111 accumulation was compared to that of the βLNNd-Lmβ1ΔLN complex at 28 nM laminin + βLNNd in panels C and D. Panel E. Surface contour of mouse laminin β1LN and adjacent LEa domain segment with attached N-linked carbohydrates (N-CHO) showing location of the mutations examined in βLNNd proteins (Pymol rendition of Protein Data Bank (PDB) 4AQS shown [35]). Polymerization-deficient mutations S68R/A, H135R, and E198K map to the LN tip and front face while R234Q maps to the LN rear face.
