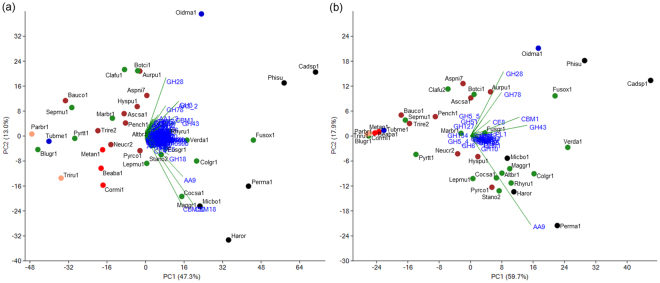
Figure 5.
Principal component analysis (PCA) of carbohydrate active enzymes (CAZymes) and plant cell wall degrading enzymes (PCWDEs) of Cadophora sp., Periconia macrospinosa, and 35 other ascomycetes including three further DSE species. (a) PCA based on CAZyme copynumbers. PC1 accounts for 47.3% of the variation and PC2 for 13%. (b) PCA based on gene copy numbers of plant cell wall degrading families. PC1 accounts for 59.7% of the variation and PC2 for 17.9%. The different fungal lifestyles are labelled in red (ap/e; animal pathogens/endophytes?), brown (sap; saprotrophs), green (plp; plant pathogens), black (dse, dark septate endophytes), blue (myc; mycorrhizal fungi), or pink (ap; animal pathogen). For complete species names and further information see Supplementary Table S5.