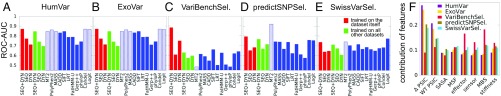
Fig. 1.
Comparison of the performance of the introduced Random Forest (RF) classifiers (SEQ+DYN, SEQ, and DYN) and existing tools for pathogenicity prediction. (A–E) AUC values derived from ROC plots (SI Appendix, Fig. S3) are presented for five datasets as indicated. The red bars refer to a 10-fold cross-validated classification on the dataset used for learning the RF classifiers; green bars refer to the RF classifiers trained on the other four datasets combined and tested on the given dataset. Solid blue bars show the AUC values from existing tools, obtained from ref. 26; dashed blue bars refer to those predictors potentially trained on the testing dataset (training bias). See SI Appendix, Fig. S2 for the results from an extended set of tools. (F) Relative contribution of eight features used in RF classifiers to pathogenicity assessment. Results for each dataset are shown in a different color. The first two features (SEQ) are residue specific, based on conservation (WT PSIC) score and its change upon mutation (ΔPSIC); the last six (DYN) are nonspecific. They account for flexibility and accessibility (SASA and MSF), allosteric properties (effector and sensor), and mechanical properties (MBS and stiffness) of sites on the 3D structure, regardless of amino acid identity. CADD, Combined Annotation Dependent Depletion; LRT, likelihood ratio test; MASS, Mutation Assessor; MT2, Mutation Taster-2.