Figure 2.
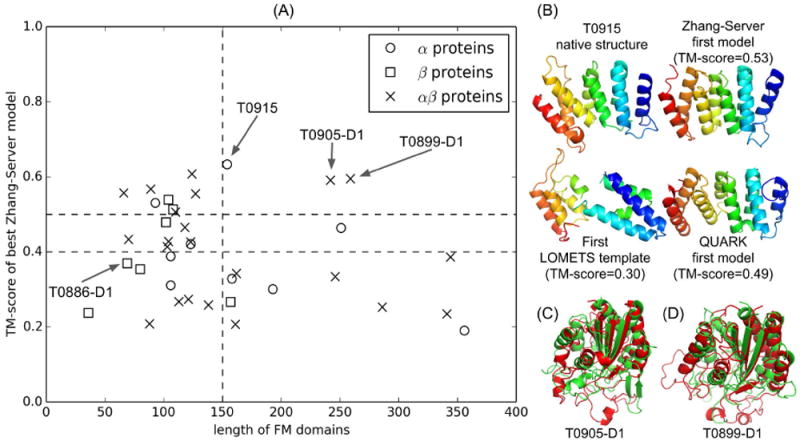
(A) TM-score of the best “Zhang-Server” model for the FM domains versus the length of each domain. The vertical dashed line represents the length cutoff of 150 residues. The horizontal dashed lines represent the TM-score cutoff of 0.4 and 0.5, respectively. Global structure folds of three medium size FM targets with length > 150, T0915, T0905-D1 and T0899-D1, are correctly predicted (TM-score > 0.5) by “Zhang-Server” as highlighted by the arrows. (B) Native structure and server models for T0915. All structures are colored in spectrum, with blue to red indicating N- to C- terminal. (C, D) “Zhang-Server” models (red) superposed onto the corresponding native structures (green) of T0905-D1 and T0899-D1, respectively.
