Figure 4.
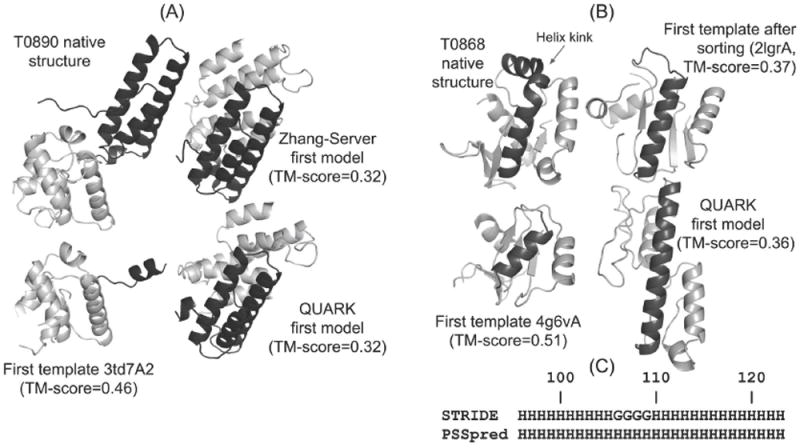
(A) The native structure, best LOMETS template, and the QUARK and Zhang-Server models for T0890, which is a two-domain protein. The domains are distinguished as per the domain assignment by CASP12 assessors, where the first domain is colored in black while the second domain is shown with grey. (B) The native structure, the template structure (before and after sorting) and the first QUARK model for T0868. Residues 96 to 123 are colored in black. (C) Secondary structure of residues 96 to 123 of T0868 assigned by STRIDE using native structure and that predicted by PSSpred. “H” stands for α-helix in STRIDE and any helix type in PSSpred. “G” stands for 3/10 helix in STRIDE.
