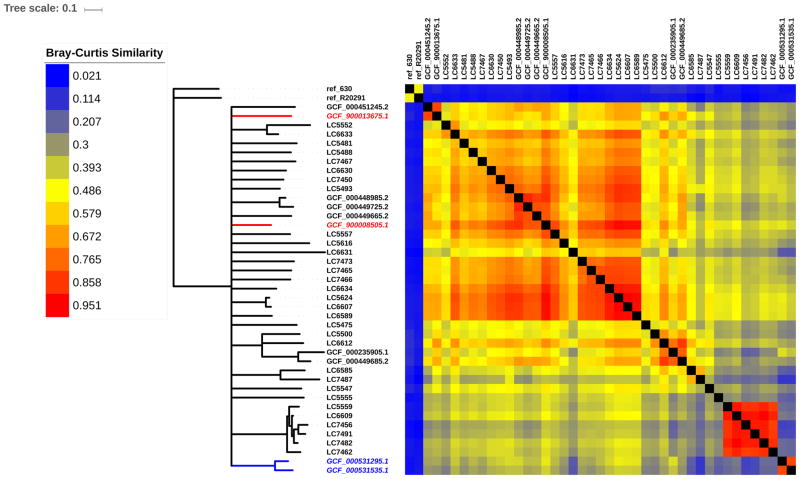
Fig 4. Relative amount of shared accessory genome content among 41 C. difficile strains monophyletic with REA group DH.
The 31 REA group DH isolates from the pediatric cohort are labeled LC and their unique identifier, and the 10 isolates from the NCBI cohort that are monophyletic with REA group DH are labeled by their NCBI GenBank assembly accession numbers. Based on review of NCBI data linked to each sequence accession number, all isolates were sequenced in the US with the exception of two sequences from Quebec, Canada (labels italicized in red font) and one sequence each from Ireland and the UK (both labels italicized in blue font). For comparison, reference strains 630 and R20291 (an epidemic BI/NAP1/027 strain) are included. Bray-Curtis distances (d) were calculated for every pairwise comparison of shared AGE content between strains. Neighbor-joining tree (left) is a consensus across 100 bootstrap resamplings of AGE distributions. Branches with support < 50 were collapsed. Heatmap (right) shows relative pairwise AGE content similarity (1 - d) between strains.