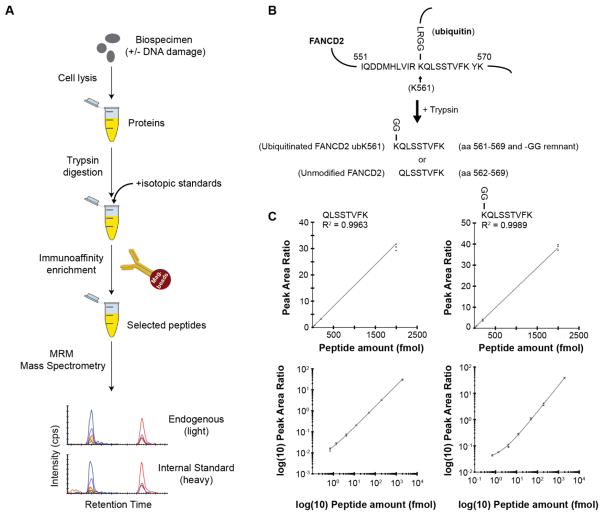
Fig. 1. Development and characterization of the multiplex assay.
(A) Description of the assay workflow: A variety of sample types can be analyzed by MRM, with previous applications demonstrated in plasma, serum, cell line lysates, and tissue specimens. Proteins are extracted from the biological sample using standard procedures amenable to downstream mass spectrometry. The protein lysates are enzymatically digested, typically using trypsin, and an HPLC-purified, stable isotope-labeled standard (SIS) peptide corresponding to each analyte is spiked-in at a known concentration to act as an internal standard. Although each pair of spiked-in SIS peptide and its endogenous analyte peptide share the same amino acid sequence (and hence the same chemical and physical properties), they are separately detected by the mass spectrometer due to the m/z difference introduced by the stable isotope label. The analyte peptides, along with their stable-isotope analogs, are enriched from the complex sample using anti-peptide antibodies. Measurement of the enriched analytes and standards is performed using quantitative mass spectrometry in a technique called multiple reaction monitoring (MRM). MRM focuses the analytical capability of the instrument on the specific analyte of interest by using a combination of mass filters in a ‘transition’ (i.e. a specific combination of precursor and fragment ions). Transitions can be cycled through sequentially to allow multiplexing. Specificity is confirmed through orthogonal biophysical properties of the analyte peptides: detection of multiple m/z transitions per peptide, chromatographic co-elution (i.e. identical retention time) of the analyte and internal standard, and identical relative abundance of transitions in the analyte and internal standard. The peak area ratio (light endogenous analyte peptide to heavy SIS peptide) is used for precise, relative quantification. Synthetic peptide standards allow the assays to be highly standardized across laboratories. (B) Design of the assay for the unmodified and monoubiquitinated FANCD2 proteoforms. Ubiquitination of FANCD2 at lysine 561 (depicted in the uppermost figure) blocks the lysine from proteolysis during trypsin digestion. This results in formation of the peptide sequence from amino acids 561–569 containing a –GG remnant on the modified lysine. Peptides containing the modified remnant and the unmodified tryptic peptide were selected for assay development. (C) Characterization of the immuno-MRM assay by response curves. Curves are plotted on linear and log(base 10) scales. Error bars are the standard deviation of technical triplicates.