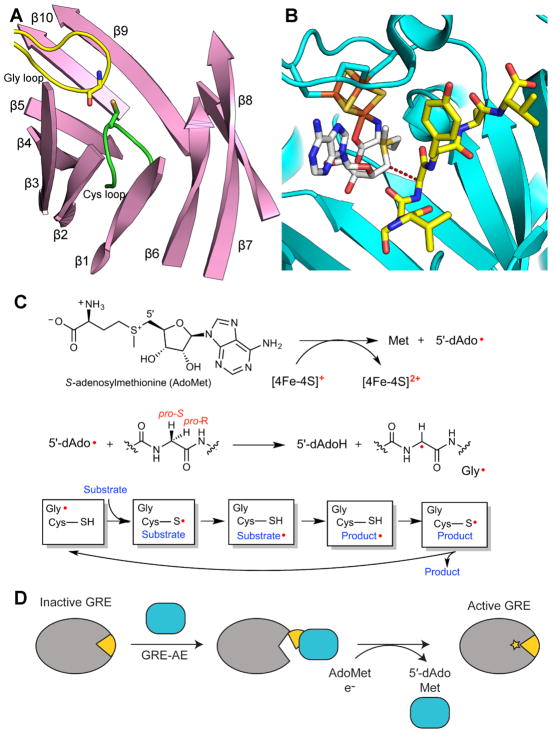
Figure 2. General overview of GRE activation and catalysis.
A. Ten β-strands surrounding the active site and two catalytic loops, the Gly (yellow) and Cys (green) loops, in the GRE PFL (PDB ID: 1H18). The α-helices and connecting loops are not shown. B. Active site of GRE activase PFL-AE (teal), showing the [4Fe-4S] cluster (orange and yellow), AdoMet (carbons in grey), and a 7-mer peptide substrate (yellow carbons) (PDB ID: 3CB8). Dashed line represents the 3.9-Å distance between the 5′ position of AdoMet and glycine residue of the 7-residue peptide, which has the same sequence as the Gly radical loop of PFL. C. A [4Fe-4S]1+ cluster reductively cleaves AdoMet to generate L-methionine and a 5′-dAdo radical species. The 5′-dAdo radical directly abstracts the pro-S hydrogen atom from the GRE glycine. Hydrogen atom transfer is thought to occur between the Gly radical and a neighboring Cys in the Cys loop to reversibly form a thiyl radical. This thiyl radical initiates catalysis. D. GRE-AE (teal) installs a Gly radical on the GRE in a reaction that is thought to require a conformational change of a region of the GRE known as the Gly radical domain (yellow). A color version of this figure is available online.