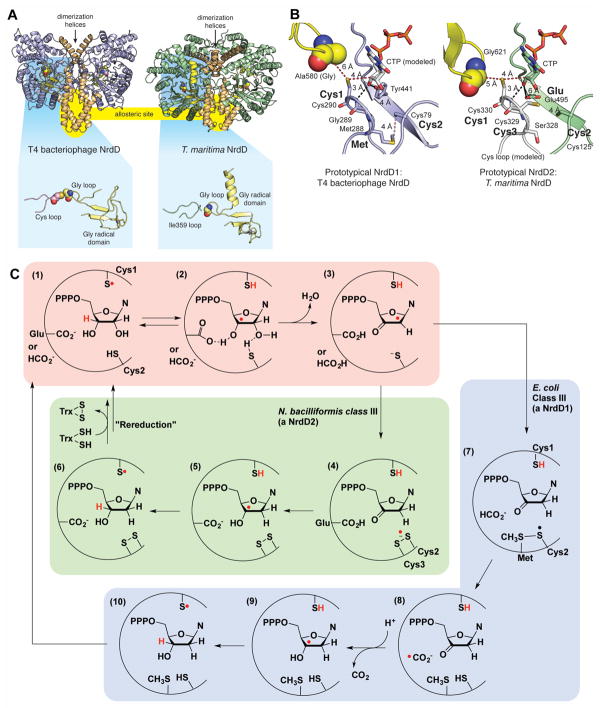
Figure 5. Structure and mechanism(s) of class III RNRs (NrdDs).
A. Two NrdD structures with helices comprising the dimeric interface shown in brown. Location of an allosteric site at the dimeric interface is highlighted in yellow. Zoomed out views of the active sites are shown highlighted against a blue background. Both structures contain a zinc (grey sphere) site in the Gly radical domain (shown in yellow), but in the T. maritima structure (PDB 4U3E), the Cys loop (pink) is missing, replaced by a different loop (Ile359 loop in green). B. Active site models that are based on the structures of a formate-utilizing NrdD1 (T4 phage NrdD) and a disulfide-utilizing NrdD2 (T. maritima NrdD). Key residues for catalysis are marked in bold. Distances are given in ångstroms. CTP in the T4NrdD structure is modeled from TmNrdD (PDB ID 4COJ). The Cys loop in TmNrdD was generated manually in PyMol through in silico mutagenesis and torsion angle adjustment to relieve clashes. C. Mechanistic proposals for NrdD1 and NrdD2 enzymes (Wei et al., 2014a, Wei et al., 2014b). See text for description. A color version of this figure is available online.