Abstract
In 2012, a new norovirus GII.4 variant (GII.4 Sydney) emerged and caused the majority of the acute gastroenteritis outbreaks in Australia, Asia, Europe, and North America. We examined the epidemiologic and molecular virologic characteristics of reported acute gastroenteritis outbreaks determined to be caused by norovirus in Taiwan from January 2012 to December 2013. A total of 253 (45.7%) of 552 reported acute gastroenteritis outbreaks tested positive for norovirus, of which 165 (65.5%) were typed as GII.4 Sydney. GII.4 Sydney outbreaks were reported from all geographic areas of Taiwan and occurred most frequently in schools (35.8%) and long-term care facilities (24.2%). Person-to-person transmission was identified in 116 (70.3%) of the outbreaks. Phylogenetic analyses of full-length ORF2 of eight specimens indicated that GII.4 Sydney strains detected in Taiwan were closely related to strains detected globally. Continued outbreak surveillance and strain typing are needed to provide information on epidemiologic and virologic trends of novel norovirus strains.
Keywords: norovirus GII.4 Sydney, epidemiology, outbreak, phylogenetic analysis
INTRODUCTION
Noroviruses are a major cause of epidemic and sporadic gastroenteritis among people of all ages, accounting for an estimated 18% of all cases worldwide [Ahmed et al., 2014]. In the United States, noroviruses cause an estimated 570–800 deaths, 56,000–70,000 hospitalizations, 400,000 emergency department visits, and 1.7–1.9 million outpatients visits annually [Hall et al., 2013a]. Norovirus-associated gastroenteritis outbreaks occur most commonly in semi-closed settings, such as hospitals, long-term care facilities, cruise ships, and schools [Glass et al., 2009]. Transmission of virus mainly occurs via the fecal–oral route, but also may occur via aerosolized vomitus and contact with fomites [Glass et al., 2009; Hall et al., 2012; Wikswo et al., 2012].
Noroviruses are small, round (27–38nm in diameter), non-enveloped, positive-sense RNA viruses belonging to the family Caliciviridae. The human norovirus genome is ~7.5 Kb in size and encodes three open reading frames (ORF1, ORF2, and ORF3). ORF1 encodes non-structural proteins, such as RNA-dependent RNA polymerase and protease. ORF2 and ORF3 encode for the major capsid protein VP1 and minor capsid protein (VP2), respectively. VP1 is further divided into the shell (S) and protruding (P) domains, with the P domain comprising the P1 and P2 subdomains [Prasad et al., 1999]. The P2 subdomain contains histo-blood group antigen (HBGA) binding sites the virus uses to bind to host cells [Tan et al., 2003, 2008].
Noroviruses are grouped into seven genogroups, and at least 40 different genotypes are recognized by capsid gene nucleic acid similarity [Vinje, 2014]. To date, viruses belonging to three genogroups (GI, GII, and GIV) are known to cause human disease [Karst et al., 2014]; genogroup II, genotype 4 (GII.4) viruses are the most common cause of norovirus gastroenteritis outbreaks worldwide [Siebenga et al., 2009; Chen and Chiu, 2012]. Over the past two decades, new GII.4 variants have emerged every 2–3 years replacing previously dominant GII.4 viruses, with GII.4 Sydney 2012 as the current predominant variant [Bennett et al., 2013; van Beek et al., 2013]. Increased norovirus activity has been associated with the emergence of several, but not all, of these variants [Glass et al., 2009; Bull and White, 2011].
To better estimate the burden of disease of norovirus outbreaks in Taiwan, we examined the epidemiologic and molecular characteristics of norovirus outbreaks reported to the Taiwan Centers for Disease Control (Taiwan CDC) from January 2012 to December 2013.
MATERIALS AND METHODS
Acute Gastroenteritis Outbreak Reporting
In Taiwan, county public health agencies are responsible for investigating and reporting suspected gastroenteritis outbreaks. Acute gastroenteritis (AGE) outbreaks, defined as ≥2 cases of diarrhea or vomiting in the same setting and time period, are reported to the Taiwan CDC through two passive surveillance systems: (i) the Notifiable Diseases Surveillance System, which maintains reports of foodborne disease outbreaks, defined as acute gastroenteritis outbreaks in which cases developed symptoms after consuming the same food, and (ii) the Symptom Surveillance System, which maintains reports of all AGE outbreaks excluding foodborne outbreaks. Data reported to these surveillance systems include case counts with demographic and clinical information, dates of illness onset and of exposure to contaminated food or infected persons, geographic location, and outbreak setting. A confirmed norovirus outbreak is defined as an AGE outbreak for which ≥1 stool specimen is positive for norovirus by laboratory testing.
Specimen Collection and Viral RNA Extraction
Based on the clinical symptoms of AGE cases, 2,428 stool specimens from 552 acute gastroenteritis outbreaks were sent to the Taiwan CDC for viral testing. All specimens were tested for norovirus and rotavirus; specimens that tested negative for these viruses were further tested for adenovirus, aichi virus, astrovirus, echovirus, picobirnavirus, and sapovirus. Specimens from 485 outbreaks also were tested for bacterial pathogens including Bacillus cereus, Salmonella spp., Staphylococcus aureus, Vibrio cholerae, and Vibrio parahaemolyticus.
Fecal samples were diluted to 10% suspension with phosphate-buffered saline and then clarified by centrifugation at 3000 × g for 15 min at 4°C. Viral RNA was extracted using the MagNA Pure Compact system (Roche Diagnostics, Indianapolis, IN) according to the manufacturer’s instructions.
Reverse Transcription Polymerase Chain Reaction (RT-PCR) to Detect Norovirus
Previously published oligonucleotide primer pairs G1SKF/R and G2SKF/R were used for the detection of norovirus GI and GII, respectively [Kojima et al., 2002]. Briefly, reverse transcription (RT) was performed at 37°C for 15 min, followed by 50°C for 1 hr using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA) and random primers (Invitrogen). The PCR products were then sequenced and genotyped using an online typing tool (http://www.rivm.nl/mpf/norovirus/typingtool) [Kroneman et al., 2011].
To further study the molecular epidemiology of the GII.4 viruses detected in this study, eight GII.4 Sydney 2012 positive samples were selected from outbreaks that occurred in 2012 (March, July, August, and September) and 2013 (February, March, April, and August) for full-length ORF2 analysis [Motomura et al., 2010]. RT was performed at 50°C for 1 hr using Superscript III reverse transcriptase (Invitrogen) and 1 µL TX30SXN (10 µM) primer, according to the manufacturer’s instructions. Polymerase chain reaction (PCR) then was performed using G2SKF-Clone and TX30SXN primers with the KOD-Plus Neo system (Toyobo, Tokyo, Japan). The PCR mix (50 µl) contained 1X Blend-TaqPlus PCR buffer, 200mM dNTP, 200nM primers and 1.25U Blend-Taq enzyme. The cycling conditions included denaturation at 94°C for 3 min, followed by 40 cycles of 94°C for 30 sec, 60°C for 30 sec, 72°C for 3 min, and a final extension at 72°C for 10 min.
Sequencing and Phylogenetic Analysis
RT-PCR products were purified by the T-Pro Gel Extraction Kit (T-pro Biotechnology, Taipei, Taiwan). Nucleotide sequence reactions were prepared with the Terminator Cycle Sequence Kit (version 3.1) and determined with the ABI 3100 Avant Sequencer (Applied Biosystems, Waltham, Massachusetts). To determine the identity of the GII.4 Sydney strains found in Taiwan with the strains detected during the same time period globally, nucleotide sequences of complete ORF2 were aligned with ClustalW using Kimura’s two parameters, and phylogenetic trees were generated from full-length ORF2 nucleotide sequences using the neighbor-joining method by MEGA 4.0 software, and bootstrap values were computed with 1,000 replication [Tamura et al., 2007]. GII.4 reference variant strains included GII.4 Den Haag 2006, GII.4 New Orleans 2009, and GII.4 Sydney 2012, as well as GII.4 strains detected in Taiwan during 2004–2012: 04-R-2 (Hunter 2004), 06-AM-11(2006a), 07-B-1(2006b), 08-F-2(2008a), 08-W-1 (Apeldoorn 2007), 09-L-4 (2009a), 09-BI-2-1(New Orleans 2009), 12-AY-1(New Orleans 2009), 12-BA-1(Sydney 2012), 12-BQ-1(Sydney 2012), 12-CD-2-4(Sydney 2012), 12-CG-2-4(Sydney 2012), 13-AS-1(Sydney 2012), 13-AV-1(Sydney 2012), 13-BE-4 (Sydney 2012) and 13-Z-2 (Sydney 2012). Nucleotide sequence data determined in this study were deposited in GenBank under the following accession numbers: 04R-2(HQ456320), 06-AM-11(KC792278), 07-B-1(HQ456329), 08-F-2 (HQ456335), 08-W-1(HQ456341), 09-L-4(HQ456343), 09-BI-2-1(HQ456346), 12-AY-1(KC792279), 12-BA-1 (KJ533132), 12-BQ-1(KC792281), 12-CD-2-4 (KC792282), 12-CG-2-4(KJ533133), 13-AS-1(KJ433968), 13-AV-1 (KJ33970), 13-BE-4(KJ51059) and 13-Z-2 (KJ533134). These data also included the following reference GII.4 variants: Sydney 2012 strains (JX459908 and JX629458), New Orleans 2009 strains (JN595867 and GU445325), Apeldoorn 2007 strains (GQ246794, AB541274, AB491291, AB445395, HQ009513 and GU270580), Yerseke 2006a strains (EF126963, GQ849126, AB447458 and EF126964), Den- Haag 2006b strains (EF126965 and EF684915), Hunter 2004 strains (DQ078814 and AY883096), Camberwell strain (AF145896, AY030098) and Lordsdale (X76716, X86557). The data also included the following GII ORF1 reference sequences: GII.e (AB434770, JF697282), GII.j (AY682552), GII.4 (EF126961, X76176), GII.12 (AB220922, AB354294, AF504671), GII.m (EU921353), GII.1 (U07611), GII.a (AB190457, AF190817), GII.g (DQ379714, GQ845370), GII.h (AB089882), GII.c (AY134748), GII.21 (AY682549, AY919139), GII.f (AY682550), GII.2 (DQ456824, X81879), GII.5 (AF397156), GII.16 (AY682551, AY772730), GII.13 (EF529741, EU921354), GII.3 (U02030, U22498), GII.22 (AB212306), GII.n(GQ856469, HM635128), GII.11 (AB126320, AY823306, AB074893), GII.18 (AY823304, AY823305), GII.8 (AB039780), GII.20 (EU275779, EU424333), GII.7 (AB039777, AB258331) and GII.6 (AB039778, JX989075). In addition to ORF2 sequencing, next generation sequencing (NGS) and 50 RACE were performed on four selected strains for complete ORF1 sequencing. The cDNA library was prepared with the NEB Next Ultra RNA Library Prep Kit for Illumina (New England Biolabs, Ipswich, Massachusetts) and then sequenced by the Illumina MiSeq 300 cycles sequence kit according to manufacturer’s protocol. NGS sequence data were analyzed using Genomics Work Bench version 7 software provided by CLC bio (Qiagen, Hilden, Germany). NGS sequence results were confirmed by long distance RT-PCR and PCR direct sequencing using the primer working method.
Data Analysis
We examined the number, etiology, and temporal trends of reported gastroenteritis outbreaks for which stool specimens were sent for laboratory testing to the Taiwan CDC from January 2012 to December 2013. Descriptive frequencies were calculated for epidemiologic information, including geographic location, outbreak setting, and mode of transmission, seasonality, and norovirus genotype.
RESULTS
Etiology of Acute Gastroenteritis Outbreaks
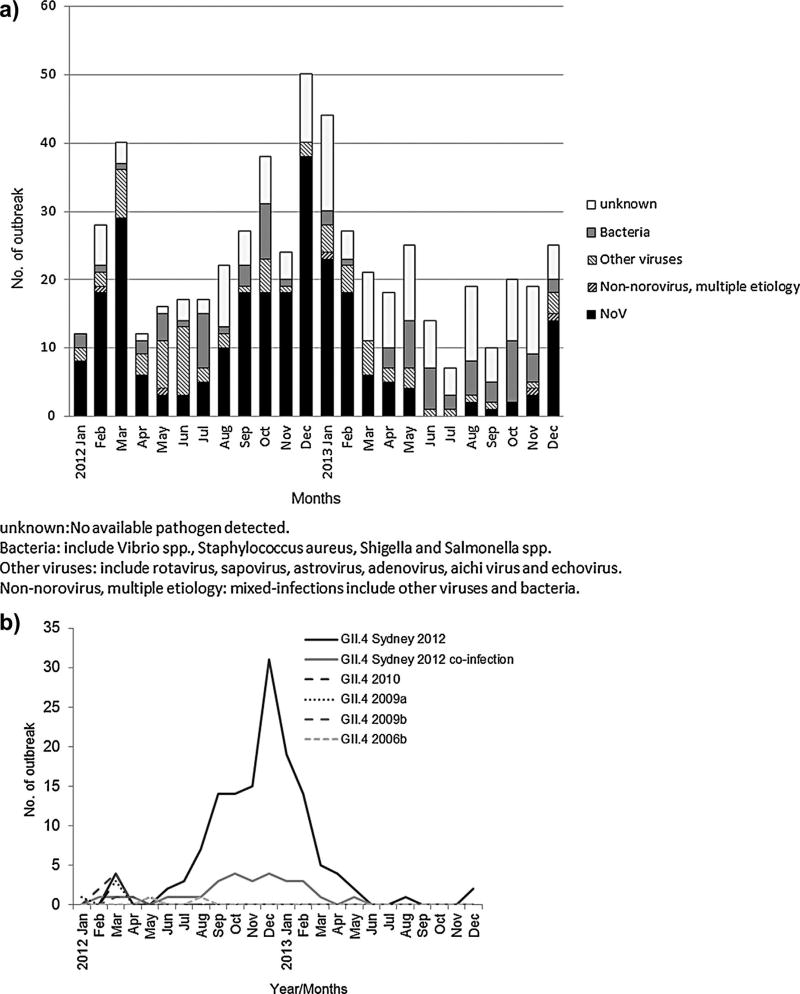
Overall, an enteric pathogen was detected in specimens from 406 (73.6%) of the 552 AGE outbreaks reported to the Taiwan CDC (Table I, Fig. 1a). Stool specimens from 253 (45.7%) outbreaks tested positive for norovirus (single and multiple etiology outbreaks), whereas specimens from 75 (13.6%) outbreaks tested positive for bacterial pathogens (i.e., Salmonella spp., Shigella spp., Staphylococcus aureus, Vibrio spp.). Other viral pathogens (i.e., rotavirus, sapovirus, astrovirus, adenovirus, aichi virus, echovirus) were detected in specimens from 70 (12.7%) outbreaks. Five (0.9%) outbreaks tested positive for both viruses and bacteria. No etiologic agent was identified in 149 (27.0%) outbreaks. Of the 253 norovirus outbreaks, 180 (71.1%) could be typed as GII.4 (0.8% GII.4 DenHaag 2006b, 2.0% GII.4 2009a, 3.2% GII.4 New Orleans, and 65.2% GII.4 Sydney), 72 (28.5%) as other GII viruses (i.e., non-GII.4), and 1 (0.4%) as GI (Table I).
TABLE I.
Etiology of Acute Gastroenteritis Outbreaks in Taiwan, January 2012–December 2013
| Pathogen | No. (%) of outbreaks (N = 552) |
|---|---|
| Norovirus (all) | 253 (45.7%) |
| By genotype, variant: | |
| GII.4 DenHaag2006bb | 2 (0.8%)a |
| GII.4 2009ac | 5 (2.0%) |
| GII.4 New Orleans 2009d | 8 (3.2%) |
| GII.4 Sydneye | 165 (65.2%) |
| GII (non-GII.4)f | 72 (28.5%) |
| GI | 1 (0.4%) |
| Non-norovirus, multiple etiologyg | 5 (0.9%) |
| Other virusesh | 70 (12.7%) |
| Bacteriai | 75 (13.6%) |
| Unknownj | 149 (27.0%) |
Percentages for norovirus genotypes are the percentage of all reported norovirus outbreaks.
Includes one outbreak for which GII.4 DenHaag2006b was identified along with bacteria.
Includes one outbreak for which GII.4 2009a was identified along with GII.12 and GI.
Includes six outbreaks for which GII.4 New Orleans 2009 was identified along with GII.4 DenHaag2006b, GII.5, GII.6, GII.12, GI.14, and bacteria.
Includes 28 outbreaks for which GII.4 Sydney2012 was identified along with GI/GII noroviruses, other viruses, or bacteria.
Includes 23 outbreaks for which other GII (GII.1, GII.2, GII.5, GII.6, GII.13, GII.17 and GII unknown genotype)was identified along with GI noroviruses, other viruses, and bacteria.
Mixed infections include other viruses and bacteria.
Other viruses include rotavirus, sapovirus, astrovirus, adenovirus, aichi virus andechovirus.
Bacteria include Vibrio spp., Staphylococcus aureus, Shigella, and Salmonella spp.
No pathogen detected.
Fig. 1.
Number of acute gastroenteritis outbreak by month of etiology in Taiwan, January 2012–December 2013. (a) All identified pathogens of outbreaks by month. (b) Norovirus GII.4 outbreaks by month. The GII.4 Sydney co-infection indicates that GII.4 Sydney was identified along with other GI/GII norovirus, viruses or bacteria.
Epidemiology of Norovirus GII.4 Sydney Outbreaks
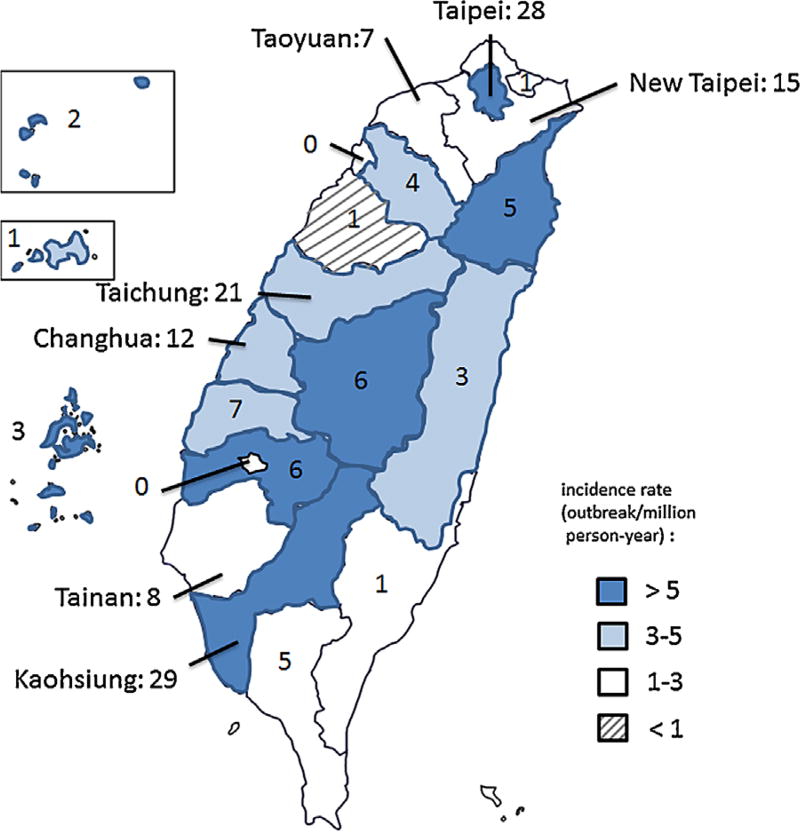
The number of reported GII.4 Sydney outbreaks peaked during the winter months with 127 (77%) of outbreaks occurring between September and February (Fig. 1b). Outbreaks were reported by all 20 counties (range: 1–29 outbreaks reported per county), with a median rate of 7.4 outbreaks per million person years per county (range: 1.8–178.2 outbreaks per million person years per county) (Fig. 2) and occurred most frequently in schools (59/165, 35.8%) and long-term care facilities (40/165, 24.2%), followed by restaurants (28/165, 17%), hospitals (21/165, Norovirus GII.4 Sydney 2012 in Taiwan 1465 12.7%), and prisons or military bases (3/165, 1.8%) (Table II). Person-to-person transmission was identified as the primary mode of transmission in 116 (70.3%) outbreaks, followed by foodborne transmission in 47 (28.5%) outbreaks (Table II). Among 422 GII.4 Sydney positive patients, 12 (2.8%) were 0–4 years of age, 133 (31.5%) were 5–20 years of age, 141 (33.4%) were 21–64 years of age, and 136 (32.2%) were ≥65 years of age. Clinical outcome data for these patients were not available.
Fig. 2.
The Number and incidence rate of norovirus GII.4 Sydney outbreaks, by county—Taiwan, January 2012–December 2013. The names of counties with >1 million in population are indicated.
TABLE II.
Number (%) of Norovirus GII.4 Sydney Outbreaks in Taiwan, by Setting and Mode of Transmission—January 2012–December 2013
| Genotype | No. (%) outbreaks (N = 165) |
|---|---|
| Setting | |
| School | 59 (35.8%) |
| Long-term care facility | 40 (24.2%) |
| Restaurant | 28 (17%) |
| Hospital | 21 (12.7%) |
| Prison/military | 3 (1.8%) |
| Other/unknown | 14 (8.5%) |
| Mode of transmission | |
| Person-to-person | 116 (70.3%) |
| Foodborne | 47 (28.5%) |
| Unknown | 2 (1.2%) |
Additional Typing of GII.4 Sydney Viruses
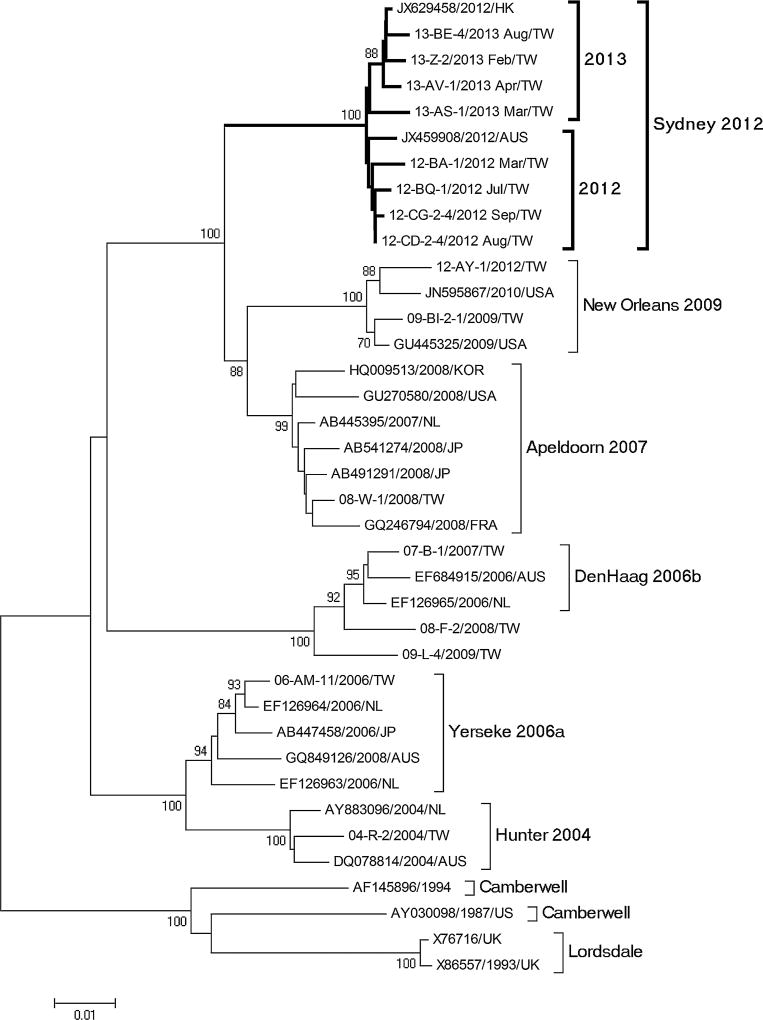
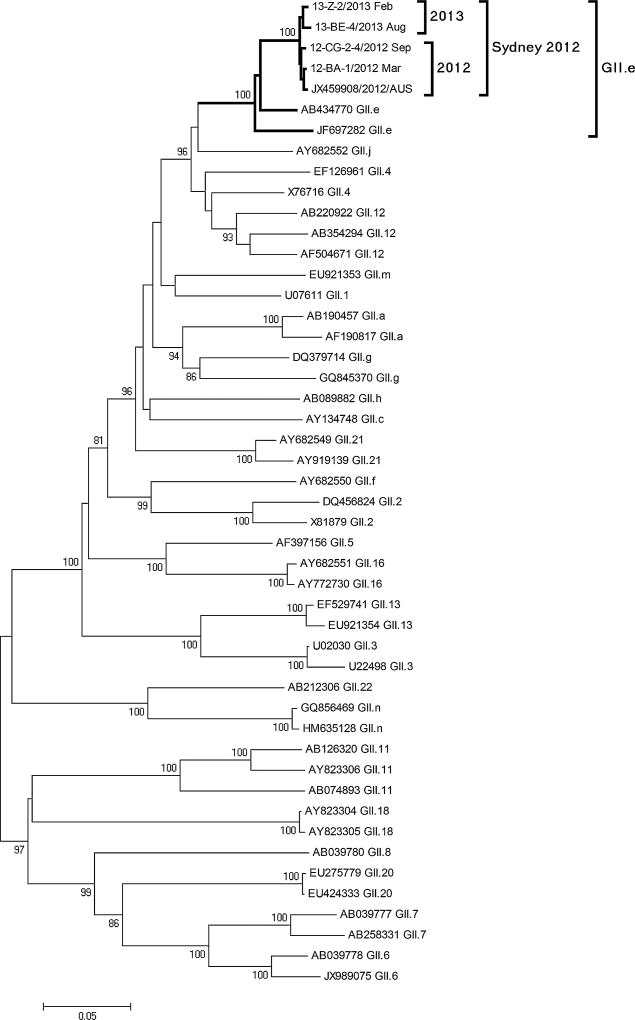
Phylogenetic analysis of full-length ORF2 showed that the 8 selected GII.4 Sydney strains had 98.7–99.4% nucleotide sequence identity with GII.4 Sydney strains detected globally (Fig. 3). Similarly, ORF1 typing showed that all GII.4 Sydney strains could be typed as GII.e identical to the prototype Sydney GII.4 strain detected in Australia in 2012 (Fig. 4).
Fig. 3.
Phylogenetic analysis of full-length norovirus GII.4 ORF2 gene. Bootstrap values of 1,000 replications are shown on the branches. Strains in boldface indicate the eight representative strains of GII.4 Sydney in Taiwan (GenBank: KC792281-KC792283, KJ433968, KJ433970, KJ451059, and KJ533132-4). Reference sequences from GenBank are named by accession number, year, and country of detection. Reference sequences from the Taiwan CDC are indicated by outbreak number, country, and collection date.
Fig. 4.
Phylogenetic analysis of partial ORF1 region of norovirus GII.4 Sydney strains in Taiwan, 2012–2013. Bootstrap values of 1,000 replications are shown on the branches. Strains in boldface indicate four representative GII.4 Sydney strains detected in Taiwan. Norovirus GII reference sequences included in the analysis were downloaded from GenBank are named by accession number and ORF1 genotype. Reference sequences from the Taiwan CDC are indicated by outbreak number, country, and collection date.
DISCUSSION
Norovirus was the most common cause of reported acute gastroenteritis outbreaks in Taiwan. GII.4 Sydney viruses caused 65.2% of reported norovirus outbreaks between January 2012 and December 2013, and we observed almost complete replacement of the previously dominant GII.4 variant (GII.4 New Orleans) since April 2012. This finding is consistent with reports worldwide [Kroneman et al., 2011; Zakikhany et al., 2012; Giammanco et al., 2013, 2014; Leshem et al., 2013; Rahman et al., 2013; Bruggink et al, 2014; Thongprachum et al., 2014], with a slight difference that no low-level co-circulation of GII.4 New Orleans viruses was found as has been reported in other countries [Fonager et al., 2013]. Our findings that GII.4 Sydney norovirus outbreaks in Taiwan predominantly occurred via person-to-person transmission are similar to what has been reported in the literature [Leshem et al., 2013]. Interestingly, schools were found as the most common setting for GII.4 Sydney outbreaks, whereas in most other studies, long-term care facilities were the predominant setting [Leshem et al., 2013]. This may be due to various biases in passive reporting by different countries, leading to preferential reporting of certain types of outbreaks. For example, in Taiwan, which had an estimated population of 23.4 million people in 2013, ~5 school-based and ~1.2 long-term care facility-based AGE outbreaks per million person years occurred during 2012–2013. In contrast, in the United States, ~0.5 school-based and ~2.4 health care facility-based (mostly nursing homes) AGE outbreaks per million person years occurred during 2009–2010 [Hall et al., 2013b].
Similar to what has been reported by others, phylogenetic analyses of full-length ORF2 and partial ORF1 sequences indicate that the GII.4 Sydney variant is closely related to more recently identified GII.4 variants and is not a product of viral recombination [Zakikhany et al., 2012].
This study had some limitations. Since the reporting of gastroenteritis outbreaks occurs through two passive surveillance systems, it is likely that not all norovirus outbreaks that occurred during January 2012–December 2013 were reported or that stool specimens for all outbreaks were sent for laboratory testing. Furthermore, the wide variability in county reporting and likely biases in which locations and settings norovirus outbreaks are more likely to be reported also suggest underreporting of these outbreaks. It is possible that the number of outbreaks reported by some counties was limited by the health department’s capacity to conduct outbreak investigations. Also, outbreaks in schools and long-term care facilities may be more likely to be reported than restaurant-related outbreaks, since schools and long-term care facilities contain more confined, monitored populations. Nevertheless, the number of outbreaks reported generally correlates with the population size of most of the counties (i.e., larger number of outbreaks reported by counties with larger populations). Exceptions were seen in Kinmen, Penghu, and Lianjiang, which are remote islands with much smaller populations, so only a few outbreaks resulted in increased incidence rates of outbreaks. Another limitation is that not all stool specimens were tested for both viral and bacterial pathogens. In some cases, once a pathogen was identified, no additional testing was conducted. Therefore, some outbreaks may have been reported as single etiology infections when they were, in fact, mixed infections; this would have led to underreporting of certain viral or bacterial pathogens. In addition, samples may not have been collected under optimal conditions for detecting bacterial pathogens. However, we believe that the general epidemiologic trends of GII.4 Sydney outbreaks were captured well given the similarity with reports from other countries.
During 2012–2013, GII.4 Sydney became the predominant cause of acute gastroenteritis outbreaks in Taiwan. Continued national outbreak surveillance combining data from two surveillance systems in Taiwan will help rapid identification of changes in virulence and genotype of the norovirus strains involved in outbreaks and could help identify if certain genotypes are more prevalent in certain outbreak settings (e.g., schools) compared to others (e.g., long-term care facilities).
Acknowledgments
We thank all county and local public health workers in Taiwan for their dedication to outbreak surveillance and reporting.
Grant sponsor: Taiwan Centers for Disease Control; Grant numbers: DOH101-DC-2016; MOHW103-CDC-C-315-00020.; Grant sponsor: Ministry of Health, Labor, and Welfare of Japan; Grant sponsor: Japan Society for the Promotion of Science (JSPS KAKENHI Grant)
Footnotes
DISCLAIMER
The findings and conclusions of this report are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention.
References
- Ahmed SM, Hall AJ, Robinson AE, Verhoef L, Premkumar P, Parashar UD, Koopmans M, Lopman BA. Global prevalence of norovirus in cases of gastroenteritis: A systematic review and meta-analysis. Lancet Infect Dis. 2014;14:725–730. doi: 10.1016/S1473-3099(14)70767-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett S, MacLean A, Miller RS, Aitken C, Gunson RN. Increased norovirus activity in Scotland in 2012 is associated with the emergence of a new norovirus GII.4 variant. Euro Surveill. 2013;18:pii:20349. [PubMed] [Google Scholar]
- Bruggink LD, Dunbar NL, Marshall JA. Emergence of GII.e as a major ORF 1 norovirus genotype. Infect Genet Evol. 2014;22:157–163. doi: 10.1016/j.meegid.2014.01.025. [DOI] [PubMed] [Google Scholar]
- Bull RA, White PA. Mechanisms of GII.4 norovirus evolution. Trends Microbiol. 2011;19:233–240. doi: 10.1016/j.tim.2011.01.002. [DOI] [PubMed] [Google Scholar]
- Chen SY, Chiu CH. Worldwide molecular epidemiology of norovirus infection. Paediatr Int Child Health. 2012;32:128–131. doi: 10.1179/2046905512Y.0000000031. [DOI] [PubMed] [Google Scholar]
- Fonager J, Hindbaek LS, Fischer TK. Rapid emergence and antigenic diversification of the norovirus 2012 Sydney variant in Denmark, October to December. Euro Surveill. 2013;18 [PubMed] [Google Scholar]
- Giammanco GM, De Grazia S, Tummolo F, Bonura F, Calderaro A, Buonavoglia A, Martella V, Medici MC. Norovirus GII.4/Sydney/2012 in Italy, winter 2012–2013. Emerg Infect Dis. 2013;19:1348–1349. doi: 10.3201/eid1908.130619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giammanco GM, De Grazia S, Terio V, Lanave G, Catella C, Bonura F, Saporito L, Medici MC, Tummolo F, Calderaro A, Banyai K, Hansman G, Martella V. Analysis of early strains of the norovirus pandemic variant GII.4 Sydney 2012 identifies mutations in adaptive sites of the capsid protein. Virology. 2014;450–451:355–358. doi: 10.1016/j.virol.2013.12.007. [DOI] [PubMed] [Google Scholar]
- Glass RI, Parashar UD, Estes MK. Norovirus gastroenteritis. N Engl J Med. 2009;361:1776–1785. doi: 10.1056/NEJMra0804575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AJ, Eisenbart VG, Etingue AL, Gould LH, Lopman BA, Parashar UD. Epidemiology of foodborne norovirus outbreaks, United States, 2001–2008. Emerg Infect Dis. 2012;18:1566–1573. doi: 10.3201/eid1810.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AJ, Lopman BA, Payne DC, Patel MM, Gastanaduy PA, Vinje J, Parashar UD. Norovirus disease in the United States. Emerg Infect Dis. 2013a;19:1198–1205. doi: 10.3201/eid1908.130465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall AJ, Wikswo ME, Manikonda K, Roberts VA, Yoder JS, Gould LH. Acute gastroenteritis surveillance through the National Outbreak Reporting System, United States. Emerg Infect Dis. 2013b;19:1305–1309. doi: 10.3201/eid1908.130482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karst SM, Wobus CE, Goodfellow IG, Green KY, Virgin HW. Advances in Norovirus Biology. Cell Host Microbe. 2014;15:668–680. doi: 10.1016/j.chom.2014.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Kageyama T, Fukushi S, Hoshino FB, Shinohara M, Uchida K, Natori K, Takeda N, Katayama K. Genogroup-specific PCR primers for detection of Norwalk-like viruses. J Virol Methods. 2002;100:107–114. doi: 10.1016/s0166-0934(01)00404-9. [DOI] [PubMed] [Google Scholar]
- Kroneman A, Vennema H, Deforche K, v d Avoort H, Penaranda S, Oberste MS, Vinje J, Koopmans M. An automated genotyping tool for enteroviruses and noroviruses. J Clin Virol. 2011;51:121–125. doi: 10.1016/j.jcv.2011.03.006. [DOI] [PubMed] [Google Scholar]
- Leshem E, Wikswo M, Barclay L, Brandt E, Storm W, Salehi E, DeSalvo T, Davis T, Saupe A, Dobbins G, Booth HA, Biggs C, Garman K, Woron AM, Parashar UD, Vinje J, Hall AJ. Effects and clinical significance of GII.4 Sydney norovirus, United States, 2012–2013. Emerg Infect Dis. 2013;19:1231–1238. doi: 10.3201/eid1908.130458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motomura K, Yokoyama M, Ode H, Nakamura H, Mori H, Kanda T, Oka T, Katayama K, Noda M, Tanaka T, Takeda N, Sato H, Norovirus Surveillance Group of J Divergent evolution of norovirus GII/4 by genome recombination from May 2006 to February 2009 in Japan. J Virol. 2010;84:8085–8097. doi: 10.1128/JVI.02125-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad BV, Hardy ME, Dokland T, Bella J, Rossmann MG, Estes MK. X-ray crystallographic structure of the Norwalk virus capsid. Science. 1999;286:287–290. doi: 10.1126/science.286.5438.287. [DOI] [PubMed] [Google Scholar]
- Rahman M, Nahar S, Afrad MH, Faruque AS, Azim T. Norovirus variant GII.4/Sydney/2012, Bangladesh. Emerg Infect Dis. 2013;19:1347–1348. doi: 10.3201/eid1908.130227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebenga JJ, Vennema H, Zheng DP, Vinje J, Lee BE, Pang XL, Ho EC, Lim W, Choudekar A, Broor S, Halperin T, Rasool NB, Hewitt J, Greening GE, Jin M, Duan ZJ, Lucero Y, O’Ryan M, Hoehne M, Schreier E, Ratcliff RM, White PA, Iritani N, Reuter G, Koopmans M. Norovirus illness is a global problem: Emergence and spread of norovirus GII.4 variants, 2001–2007. J Infect Dis. 2009;200:2001–2007. doi: 10.1086/605127. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4 Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol Biol Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- Tan M, Huang P, Meller J, Zhong W, Farkas T, Jiang X. Mutations within the P2 domain of norovirus capsid affect binding to human histo-blood group antigens: Evidence for a binding pocket. J Virol. 2003;77:12562–12571. doi: 10.1128/JVI.77.23.12562-12571.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Xia M, Cao S, Huang P, Farkas T, Meller J, Hegde RS, Li X, Rao Z, Jiang X. Elucidation of strain-specific interaction of a GII-4 norovirus with HBGA receptors by site-directed mutagenesis study. Virology. 2008;379:324–334. doi: 10.1016/j.virol.2008.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thongprachum A, Chan-it W, Khamrin P, Saparpakorn P, Okitsu S, Takanashi S, Mizuguchi M, Hayakawa S, Maneekarn N, Ushijima H. Molecular epidemiology of norovirus associated with gastroenteritis and emergence of norovirus GII.4 variant 2012 in Japanese pediatric patients. Infect Genet Evol. 2014;23:65–73. doi: 10.1016/j.meegid.2014.01.030. [DOI] [PubMed] [Google Scholar]
- van Beek J, Ambert-Balay K, Botteldoorn N, Eden JS, Fonager J, Hewitt J, Iritani N, Kroneman A, Vennema H, Vinje J, White PA, Koopmans M, NoroNet Indications for worldwide increased norovirus activity associated with emergence of a new variant of genotype II.4, late 2012. Euro Surveill. 2013;18:8–9. [PubMed] [Google Scholar]
- Vinje J. Advances in laboratory methods for detection and typing of norovirus. J Clin Microbiol. 2015;53:373–381. doi: 10.1128/JCM.01535-14. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wikswo ME, Hall AJ, Centers for Disease C, Prevention Outbreaks of acute gastroenteritis transmitted by person-to-person contact–United States, 2009–2010. MMWR Surveill Summ. 2012;61:1–12. [PubMed] [Google Scholar]
- Zakikhany K, Allen DJ, Brown D, Iturriza-Gomara M. Molecular evolution of GII-4 Norovirus strains. PLoS One. 2012;7:e41625. doi: 10.1371/journal.pone.0041625. [DOI] [PMC free article] [PubMed] [Google Scholar]