Figure 5. Lysosomal damage promotes interactions between Gal8 and the amino acid and cholesterol sensor SLC38A9.
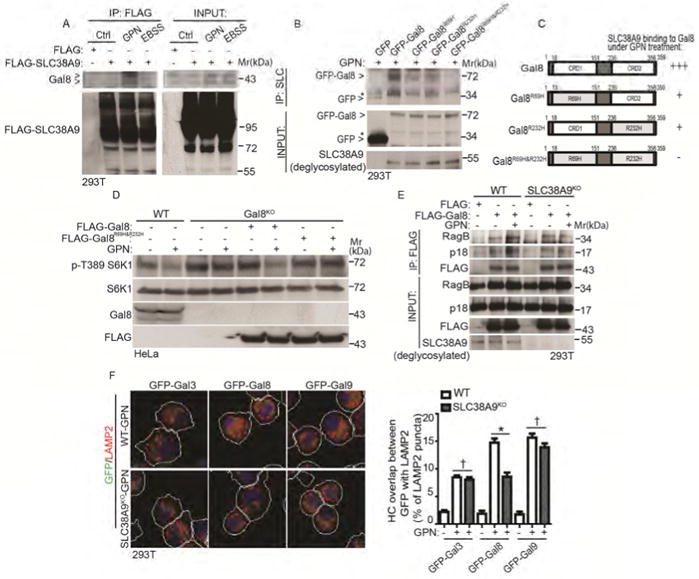
(A) Cells expressing FLAG-SLC38A9 were treated with 100 μM GPN in full medium or starved in EBSS for 1 h. Cell lysates were subjected to anti-FLAG immunoprecipitation and immunoblotted for endogenous Gal8. Control (Ctrl), untreated cells. Note: SLC38A9 is heavily glycosylated protein with smear pattern in immunoblots. (B) Co-IP analysis of interactions between SLC38A9 and Gal8. Cells expressing GFP-tagged Gal8 or glycan recognition-mutant forms of Gal8 (individual R69H, R232H or double/combined R69H & R232H; see panel C) were treated with 100 μM GPN for 1 h in full medium. IP, anti-SLC38A9 (SLC) antibody; immunoblotting with anti-GFP. *, non-specific bands. Note: input SLC3A89 was deglycosylated with PNGase F. (C) Schematic diagram of Gal8 domains (CRD and CRD2, carbohydrate recognition domains 1 and 2) and summary of interactions between SLC38A9 and Gal8. +++, strong; +, weak; -, not detectable. (D) mTOR activity in wild type (WT) and Gal8-knockout (Gal8KO) HeLa cells. Gal8KO cells transfected wtih FLAG-tagged Gal8 or double glycan (R69H & R232H) recognition-mutant form of FLAG-Gal8R69H&R232H were treated with 100 μM GPN for 1 h in full medium. mTOR activity was monitored by S6K1 (T389) phosphorylation. (E) Co-IP analysis of Gal8 with RagB or p18 in response to GPN treatment. Wild type (WT) or SLC38A9-knockout (SLC38A9KO) cells transfected with FLAG vector or FLAG-Gal8 were treated with 100 μM GPN in full medium for 1 h. IP, anti-FLAG; immunoblot, antibodies against RagB and p18. (F) HC quantification of overlaps between galectins and LAMP2 in response to GPN treatment. Wild type (WT) and SLC38A9 knockout (SLC38A9KO) HEK293T cells expressing GFP-galectin fusions were treated with 100 μM GPN in full medium for 1 h. White masks, automatically defined cell boundaries (primary objects); red and green masks, computer-identified LAMP2 and GFP-galectin profiles (target objects). HC data, means □ SEM; n ≥ 3 (each experiment: 500 valid primary objects/cells per well, ≥ 5 wells/sample). † p ≥ 0.05 (not significant), *p < 0.05, ANOVA. See also Figure S5.
