Figure 6. SLC38A9 is required for mTOR reactivation during recovery from lysosomal damage.
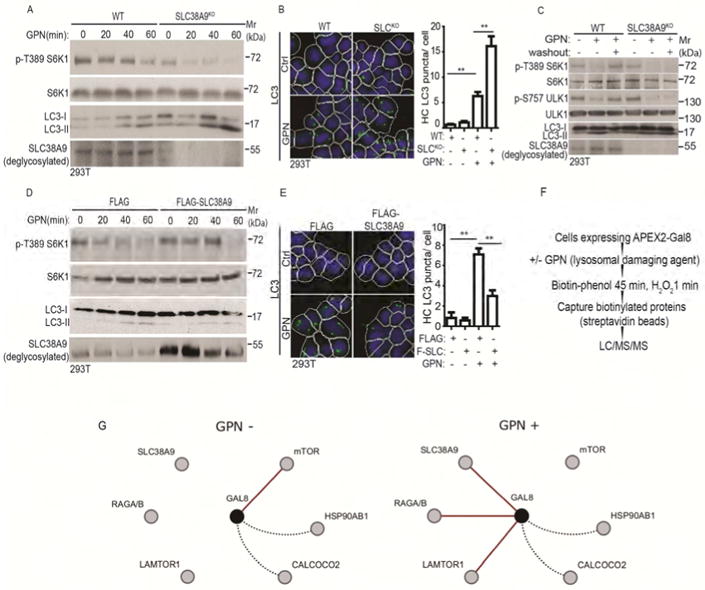
(A) mTOR activity (immunoblots; S6K1 phosphorylation at T389) and autophagy induction (LC3 immunoblots) in WT (wild type) and SLC38A9 knockout (SLC38A9KO) HEK293T cells treated with 100 μM GPN in full medium (time course). (B) HC analysis of autophagy induction (endogenous LC3 puncta) in WT and SLC38A9KO cells treated with 100 μM GPN in full medium for 30 min.. Control (Ctrl), untreated cells. White masks, algorithm-defined cell boundaries (primary objects); green masks, computer-identified LC3 puncta (target objects). (C) mTOR activity recovery and autophagy inhibition in SLC38A9KO cells after GPN washout. WT and SLC38A9KO HEK293T cells were treated with 100 μM GPN for 1 h followed by 1 h washout in full medium. mTOR activity and autophagy were monitored as in A. (D) mTOR activity and autophagy induction (monitored as in A) cells overexpressing FLAG-SLC38A9 or FLAG (vector control) treated with 100 μM GPN in full medium (time course). (E) HC analysis of autophagy induction in SLC38A9-overexpressing cells treated with GPN. FLAG and FLAG-SLC38A9 (F-SLC) expressing HEK293T cells were treated with 100 μM GPN in full medium for 30 min, and LC3 puncta were quantified by HC. Ctrl, no GPN. Masks, as in B. Data (B and E), means □ SEM, n ≥ 3 independent experiments (500 primary objects counted per well; ≥ 5 wells/sample per each experiment), **p < 0.01, ANOVA. (F) Schematic, strategy for APEX2-Gal8 LC-MS/MS proteomic analysis (see STAR methods). (G) Cytoscape depiction of dynamic changes in protein interactions/proximity relative to Gal8 in response to lysosomal damage caused by exposure to GPN (GPN+), based on protemic data in Table S1. Red lines, key changes in interactions/proximities (using > 100-fold change in precursor peak intensities as a cutoff) observed in each of the three complete biological replicates of HEK293T cells transfected with pJJiaDEST-APEX2-Gal8 subjected to separate LC-MS/MS analyses. Dotted black lines, examples of proteins identified in the LC/MS/MS analysis that did not display changes when comparing control (full medium without GPN; GPN-) vs. lysosomal damage (full medium with 100 μM GPN for 1 h; GPN+) (see Table S1). The MS/MS proteomic data have been deposited at MassIVE, ID MSV000081788 and linked to ProteomeXchange accession ID PXD008390). See also Figure S5 and Table S1.
