Figure 2.
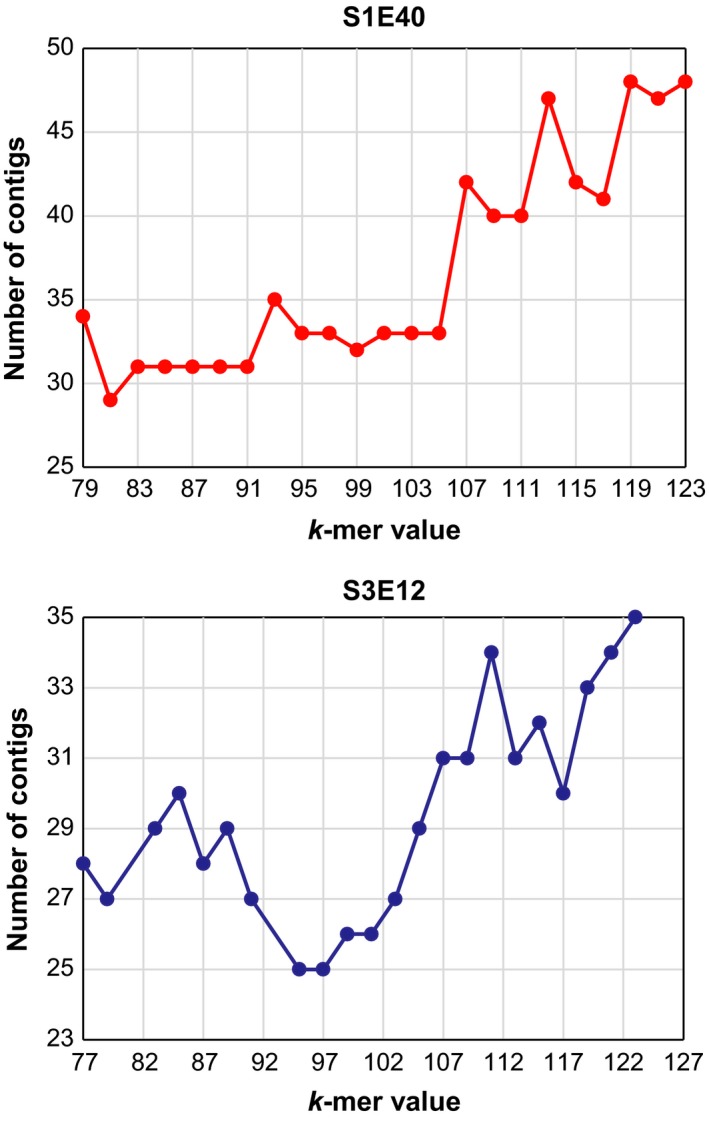
Line plots showing the influence of k‐mer values on the number of contigs obtained using ABySS assembler on Illumina MiSeq data sets for Pseudomonas sp. strains S1E40 (a) and S3E12 (b). The best k‐mer values (fewest number of contigs) for Pseudomonas strains S1E40 and S3E12 were 85 and 95, respectively
