Abstract
The interactions of two highly positively charged short peptide sequences with negatively charged lipid bilayers were explored by fluorescence binding assays and all-atom molecular dynamics simulations. The bilayers consisted of mixtures of phosphatidylglycerol (PG) and phosphatidylcholine (PC) lipids as well as a fluorescence probe that was sensitive to the interfacial potential. The first peptide contained nine arginine repeats (Arg9), and the second one had nine lysine repeats (Lys9). The experimentally determined apparent dissociation constants and Hill cooperativity coefficients demonstrated that the Arg9 peptides exhibited weakly anticooperative binding behavior at the bilayer interface at lower PG concentrations, but this anticooperative effect vanished once the bilayers contained at least 20 mol % PG. By contrast, Lys9 peptides showed strongly anticooperative binding behavior at all PG concentrations, and the dissociation constants with Lys9 were approximately 2 orders of magnitude higher than with Arg9. Moreover, only arginine-rich peptides could bind to the phospholipid bilayers containing just PC lipids. These results along with the corresponding molecular dynamics simulations suggested two important distinctions between the behavior of Arg9 and Lys9 that led to these striking differences in binding and cooperativity. First, the interactions of the guanidinium moieties on the Arg side chains with the phospholipid head groups were stronger than for the amino group. This helped facilitate stronger Arg9 binding at all PG concentrations that were tested. However, at PG concentrations of 20 mol % or greater, the Arg9 peptides came into sufficiently close proximity with each other so that favorable like-charge pairing between the guanidinium moieties could just offset the long-range electrostatic repulsions. This led to Arg9 aggregation at the bilayer surface. By contrast, Lys9 molecules experienced electrostatic repulsion from each other at all PG concentrations. These insights may help explain the propensity for cell penetrating peptides containing arginine to more effectively cross cell membranes in comparison with lysine-rich peptides.
Introduction
Many proteins and peptides contain amino acid sequences rich in arginine and lysine residues that can interact strongly with negatively charged phospholipids such as phosphatidylglycerol (PG). For example, antimicrobial peptides (AMPs) excreted from eukaryotic cells exploit electrostatic interactions with charged bacterial membranes in order to lyse them.(1) This strategy exploits the preferential interactions of these short, positively charged peptide sequences with their negatively charged bacterial membrane targets compared with zwitterionic eukaryotic membranes.(2, 3) Another class of short peptides that contain positively charged moieties is cell-penetrating peptides (CPPs), which help transport molecular cargo across membranes.(4) It is known that there is great diversity in the putative modes of operation for AMPs and CPPs with membranes,(5–8) and the mechanisms of action are still a matter of considerable debate.(9)Significantly, the efficacy of such peptides is known to depend on whether they contain multiple arginine rather than lysine residues in their sequences. In fact, peptides containing just lysine residues are not believed to penetrate the bilayer nearly as effectively as those containing arginine, although even peptides containing lysine residues are known to have both electrostatic and hydrophobic interactions with membrane lipids.(10) As such, we wished to interrogate the differences in the interactions of simple arginine and lysine sequences with PG-rich membranes.
Although the initial interactions of CPPs with plasma membranes are driven by electrostatics, the discovery that arginine residues enhance internalization over lysine suggests that the guanidinium moiety plays a crucial role in this process. Indeed, guanidinium is capable of forming bidentate hydrogen bonds with negatively charged species in the membrane, such as phosphate groups, which reduces the polarity of the guanidinium group and aids in membrane internalization.(11) By contrast, lysine moieties contain a positively charged amino group that forms only single hydrogen bonds with anionic membrane components. Additionally, the electrostatic interactions between arginine residues are attenuated by bidentate counterion scavenging.(12)
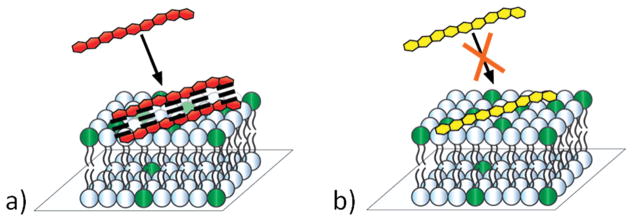
In the experiments reported below, thermodynamic binding data was obtained for Lys9 and Arg9peptides with supported lipid bilayers containing 0–30 mol % 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoglycerol (POPG). Incorporation of 0.5 mol % of a pH-sensitive fluorophore into the bilayer enabled the association of unlabeled peptides to be monitored.(13) The results demonstrated that Arg9 interacted with PG-containing membranes with a KD value that was roughly 2 orders of magnitude lower than for Lys9. This result stems, in part, from the fact that Arg9 can bind more tightly to lipid head groups than Lys9. Moreover, the lysine-containing peptides showed strongly anticooperative binding behavior, while the anticooperative effects with Arg9 were much weaker. In fact, at sufficiently high POPG concentrations, no anticooperative binding was seen for the arginine-rich sequences. Corresponding molecular dynamics simulations confirmed these findings. Such results suggest that short-range attractive forces between Arg9 molecules are roughly as strong as long-range electrostatic repulsions when these peptides come within close enough proximity. This should be due to favorable guanidinium ion pair formation over short distances.(14–17) This is quite different from lysine, where electrostatic repulsions for the amino group interactions appear to dominate at all length scales. Figure 1a illustrates the interactions of Arg9with supported lipid bilayers containing POPG, while Figure 1b shows the results found with Lys9.
Figure 1.
Schematic diagram for Arg9 (red) and Lys9 (yellow) interacting with a supported lipid bilayer containing negatively charged phosphatidylglycerol lipids (dark green). (a) Arg9 displays much less pronounced anticooperative binding behavior, which is consistent with favorable short-range interactions (dashed lines) between the guanidinium moieties on the peptide, whereas (b) Lys9 experiences strong anticooperative binding effects.
Materials and Methods
Preparation of Small Unilamellar Vesicles
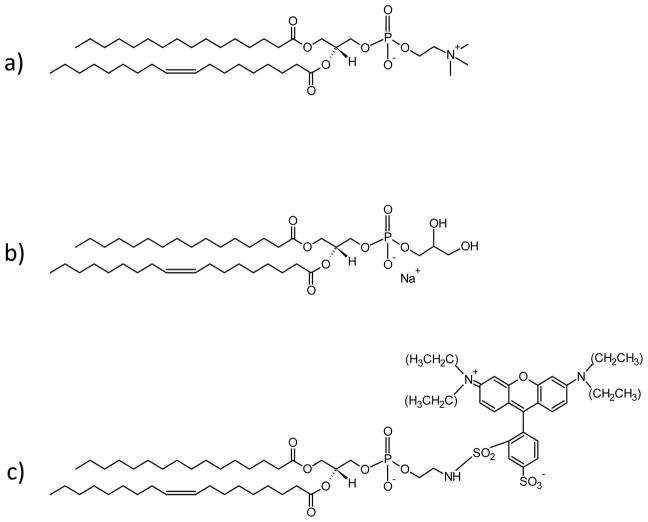
In a first step, the desired phospholipids were mixed in chloroform and then dried by purging with nitrogen gas. The mixtures contained varying amounts of 1-palmitoyl-2-oleoyl-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (POPG) and 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC) as well as 0.5 mol % of the pH-sensitive ortho isomer of rhodamine B (lissamine rhodamine B sulfonyl chloride, ortho isomer) conjugated to the free amine of POPE (1-hexadecanoyl-2-(9Z-octadecenoyl)-sn-glycero-3-phosphoethanolamine). The mixtures were desiccated under vacuum for 3 h before being reconstituted in 10 mM phosphate-buffered saline (PBS) containing 150 mM NaCl to a concentration of 0.5 mg/mL. After the lipids were fully dissolved in buffer solution, they were subjected to 10 freeze–thaw cycles by successive immersion in liquid nitrogen and warm water. This was followed by at least seven extrusions using a Lipex extruder (Northern Lipids Inc., Vancouver, Canada) through two stacked polycarbonate filters (Whatman) with 100 nm hole diameters. The size of the extruded vesicles was determined by dynamic light scattering (Brookhaven Instruments 90Plus Particle Size Analyzer) with mean sizes falling between 90 and 100 nm and a polydispersity index of 0.1–0.2. Vesicle solutions were stored at 4 °C until use. The structures of the phospholipids employed in these studies are shown in Figure 2.
Figure 2.
Structures of (a) POPC, (b) POPG, and (c) ortho-rhodamine B POPE.
Preparation of Peptide Sequences
H2N-KKKKKKKKK-NH2 (Lys9) and H2N-RRRRRRRRR-NH2 (Arg9) were synthesized by traditional Fmoc solid-phase peptide synthesis using a Rink amide MBHA resin (Novabiochem). The nascently formed Lys9 and Arg9 were purified by reversed-phase C18 HPLC, and their masses were confirmed by matrix-assisted laser desorption ionization–time-of-flight (MALDI-TOF) mass spectrometry. A detailed description of the synthetic procedures is provided in the Supporting Information.
Preparation of Glass Substrates
The glass substrates used for supporting fluid lipid bilayers were prepared from Fischer glass coverslips (24 × 40 mm2, No. 1.5) by first boiling in a 1:6 solution of 7× cleaning solution (MP Biomedicals, Solon, OH) and purified water (Barnstead Nanopure) for at least 1 h. This was followed by rinsing profusely with purified water before drying thoroughly with nitrogen gas. The coverslips were then annealed for 5 h at 520 °C before being stored until use.
Fabrication of Flow Cells
A 1.5 μm thick sheet of polydimethylsiloxane polymer (PDMS) was produced by curing the polymer between two hydrophobized glass slides. A hole was cut into the center of the PDMS sheet, and a well was made by pressing it onto a clean glass coverslip. A PDMS stamp, which was cured over a seven-channel microfluidic pattern, was incubated in a 1 mg/mL bovine fibrinogen (MP Biomedicals, Solon, OH) solution and then pressed onto the glass coverslip in the well for 5 min before being removed. This left a patterned bovine fibrinogen imprint on the glass. Vesicles were then deposited in the well and incubated for 10 min before the excess was rinsed away with copious amounts of water. Continuous supported bilayers only formed in the regions lacking the bovine fibrinogen pattern. A second sheet of PDMS with an inlet and an outlet hole was then placed on top of the well, and the entire structure was held together by aluminum plates with two clamps to make a flow cell. Finally, fresh solution was flowed through the device as experiments were performed.
Flow-Cell and pH Titration Curves
In a first set of experiments, patterned bilayers were prepared by microcontact patterning with a PDMS stamp.(18, 19) These bilayers contained varying concentrations of POPG in POPC and were placed in the flow cell described in the previous section. A fluorescence micrograph of the corresponding bilayer pattern is shown in Figure S1. Next, solutions of 10 mM PBS containing 150 mM NaCl were adjusted to specific pH values and introduced into the flow cell. Fluorescence images were taken until the intensity stabilized. These values were used to form titration curves of the pH-sensitive dye as a function of POPG concentration in the bilayers (Figure S2). Apparent pKa values were extracted from the titration curves as described in the Supporting Information.
pH Modulation Sensing
Monitoring Arg9 and Lys9 binding to supported lipid bilayers was performed via pH-modulation sensing,(13) which affords the ability to investigate the interactions of small basic peptides with negatively charged phospholipid membranes under real-time, continuous flow conditions without labeling the peptides. To perform these experiments, ortho-rhodamine B conjugated POPE was synthesized as described previously(20) and employed as a sensor in supported lipid bilayers that contained varying concentrations of POPG. When Lys9 or Arg9 adsorbed to the bilayer surface, they caused an increase in the interfacial potential because of their positive charge. This in turn caused the fluorescence response from the pH-sensitive dye to attenuate (Figure 3). Bilayers containing 0.5 mol % ortho-rhodamine B POPE and varying amounts of POPG were deposited on annealed glass slides within flow chambers. PBS buffer (10 mM) with 150 mM NaCl was flowed through the flow cell until the fluorescence intensity from the bilayer stabilized. Following this, increasing concentrations of peptide were introduced into the flow cell, and fluorescence micrographs were taken every 5 min. After the fluorescence stabilized for a given peptide concentration, a higher concentration of peptide was introduced. A 10× air (N.A. = 0.45) objective and a Texas Red filter set (Chroma Technology, Bellows Falls, VT) were used with a Lumen 200 (Prior Scientific) light source to record fluorescence data on an inverted Nikon Eclipse Ti-U fluorescence microscope (Tokyo, Japan) equipped with a ProEM 1024 CCD camera (Princeton Instruments). MetaMorph software (Version 7.7.0.0, Universal Imaging) was employed for fluorescence data analysis.
Figure 3.
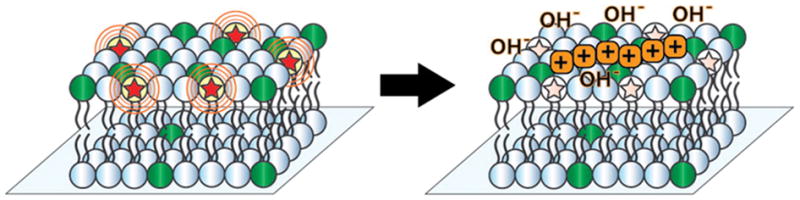
Supported lipid bilayer containing pH-sensitive dye (stars) and POPG (dark green) is exposed to a positively charged peptide (orange), thereby recruiting hydroxide ions to the membrane surface and deprotonating the fluorescent reporter.
Peptide Binding Assays
Fluorescence images were taken of supported lipid bilayers in the flow cell devices every 5 min to measure the association of Arg9 or Lys9 peptides with the bilayers. PBS buffer (10 mM) with 150 mM NaCl at pH 6.8 (or 6.4 with 5 and 0 mol % POPG) was flowed until the fluorescence intensity stabilized. Peptide at the lowest concentration was then introduced, and the fluorescence was again monitored until it stabilized. Figure S3 shows the normalized fluorescence intensity as a function of time as increasing concentrations of Lys9 are added to a bilayer containing 30 mol % POPG. It took approximately 15 min for the fluorescence to stabilize at the lowest peptide concentration, but the stabilization time required was shorter at higher peptide concentrations, as expected. From the data, it was possible to plot the change in fluorescence intensity (ΔF) normalized to the maximum change in fluorescence intensity at a saturating concentration of peptide (ΔFmax) as a function of added peptide concentration. Fitting these plots to isotherms allowed for the extraction of KD values for the peptides. The measurements were performed for each POPG concentration in the bilayer.
Total Internal Reflection Fluorescence Microscopy
Total internal reflection fluorescence microscopy (TIRFM) was performed to discriminate between peptides in solution and those bound to the surface of pure POPC bilayers.(21) 5-Carboxytetramethylrhodamine (TAMRA) labeled on the N-terminus of nonaarginine (Arg9) and nonalysine (Lys9) were purchased from Genscript. Epifluorescence microscopy (E800 fluorescence microscope, Nikon) was used to obtain fluorescent images of microchannels containing the supported membranes and peptides with a 4× objective. The microchannels used in these experiments were fabricated using standard procedures described in detail in a previous publication.(22) An 8 mW 532 nm solid-state laser beam (Dragon Lasers, China) was reflected off the sample surface through a dove prism, which through the use of immersion oil was optically coupled to the planar coverslip surface. This generated an evanescent wave at the interface which, under the conditions employed, decayed exponentially to its 1/e value within 70 nm of the surface.(22–25) This allowed measurements of the fluorescence of peptides bound to the supported lipid bilayer as opposed to peptides in bulk solution above the bilayer surface. A Photometrics Sensys CCD camera was used to capture all images, which were stored using Metamorph software from Universal Imaging Corp., and further processed using OriginLab.
Simulation Details
Molecular dynamics (MD) simulations were performed in aqueous solutions of Arg9 and Lys9 with a free N-terminus and an amide group capped C-terminus, which matched the peptides used in the experiments. These peptides were investigated in the presence of POPC and mixed POPC/POPG bilayers. The simulations were run with 150 mM NaCl as well as with 6 or 12 peptides to study the influence of concentration on binding. To maintain electroneutrality, an appropriate number of chloride counterions was added to the system. Bilayers containing 128 POPC molecules were constructed such that 64 individual POPC lipids were placed on an 8 × 8 grid, thus forming one leaflet of the bilayer. An identical procedure was used for the second bilayer leaflet. In the case of mixed POPC/POPG bilayers, containing 90 POPC (70.3%) + 38 POPG (29.7%) molecules, 19 POPC lipids were randomly replaced in each of the leaflets with the same number of POPG molecules. Equilibration of lipid bilayers was performed for at least 20 ns for the POPC bilayer and 100 ns for the POPC/POPG bilayer until constant area per lipid was achieved. Oligopeptides, together with 150 mM NaCl solution and the appropriate number of chloride counterions, were placed in a unit cell with the size of ca. 6.5 × 6.5 × 13.0 nm3 and solvated by >12000 water molecules as described with the TIP3P water model.(26) The oligopeptides were simulated with the AMBER99SB force field,(27) whereas the POPC and POPG lipid molecules were simulated using the SLipids force field.(28–30)
The system was built in a way that lipid bilayers spanned the xy plane while the z coordinate was normal to the bilayer. 3D periodic boundary conditions were employed with long-range electrostatic interactions beyond the nonbonded cutoff of 1 nm. This was accounted for using the particle-mesh Ewald procedure(31) with a Fourier spacing of 1.2 nm. The real space Coulomb interactions were cut off at 1.0 nm, while van der Waals interactions were cut off at 1.4 nm. Semi-isotropic pressure coupling employing the Parrinello–Rahman algorithm(32) was used independently in the directions parallel and perpendicular to the bilayer normal. The pressure was set to 1 bar with a coupling constant of 10 ps–1. The temperature was set to 310 K and controlled with the Nose–Hoover thermostat(33) independently for the lipid, peptide, and water subsystems, with a coupling constant of 0.5 ps–1. All bonds within the lipids and peptides were constrained using the LINCS algorithm,(34) whereas bonds in water were kept constant using the SETTLE method.(35) Equations of motion were integrated using the leapfrog algorithm with a time step of 2 fs. After equilibration of systems for at least 20 ns, a subsequent 400 ns simulation time was used for analysis. All MD simulations were performed using the GROMACS program package, version 4.5.4.(36)
Results
Arg9 and Lys9 Binding Studies with POPC
In a first set of experiments, the interactions of Arg9 were explored with supported lipid bilayers containing POPC (Figure 4a). To begin an experiment, 10 mM PBS buffer with 150 mM NaCl at a pH of 6.4 was continuously flowed over the bilayers, and fluorescence images were captured every 5 min until stabilization of the fluorescence signal intensity was achieved. At this point, the original buffer solution was exchanged for one containing a fixed concentration of Arg9, and the process was repeated over a peptide concentration range of 0–100 μM. An example plot of the fluorescence micrograph data is provided in the Figure S3. Figure 4a shows the change in fluorescence intensity (ΔF) for this system with respect to the maximum decrease that can be extrapolated from a saturation concentration of the peptide (ΔFmax). The error bars represent standard deviations from at least three measurements. As can be seen, the fluorescence decreased after introducing lower concentrations of Arg9 before leveling off at higher peptide concentrations.
Figure 4.
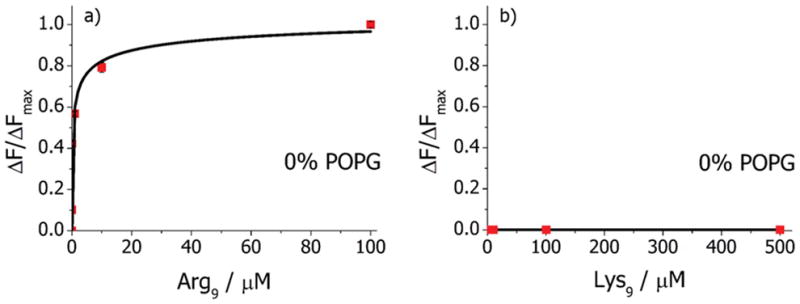
Binding isotherm on POPC bilayers for (a) Arg9 and (b) Lys9 in 10 mM PBS with 150 mM NaCl at pH 6.4.
It is possible to fit a binding isotherm to the data in Figure 4a. It should be noted that the data do not fit very well to a simple Langmuir isotherm. Instead, the Hill–Waud (eq 1) binding model was employed, which takes into account cooperativity in the binding process:
| (1) |
where ΔF/ΔFmax is the normalized measured fluorescence intensity value due to the equilibrium binding of peptide to the bilayer surface, ΔFmax is the maximum change in the fluorescence intensity at a peptide concentration that would saturate the bilayer surface, [P] is the bulk peptide concentration, KD is the apparent dissociation constant, and n is the Hill coefficient of cooperativity. The Hill–Waud equation reduces to a simple Langmuir isotherm for n = 1 (the noncooperative case). For n > 1, the binding behavior is defined as cooperative, but it is anticooperative for n < 1. The fitted data yielded a KD value of 890 nM with n = 0.39. The low value of n is expected because the first peptide interacts with an essentially uncharged membrane, but subsequent peptides need to bind to a surface with a positive interfacial potential that increases with peptide loading. A positive surface potential will electrostatically repel the positively charged peptides from the double layer above the bilayer surface, leading to the observed anticooperative binding effects.
Next, a similar set of experiments as those described above was repeated with Lys9 in place of Arg9. In this case, no peptide was found to associate with the bilayer surface as there was no modulation in fluorescence intensity at peptide concentrations up to 500 μM (Figure 4b). This result is striking as it indicates that simply changing the positive charge on the side chain from a guanidinium moiety to an amino group has a dramatic effect on the peptide interactions with phosphatidylcholine lipids.
It should be noted that the presence of ortho-rhodamine B dye in the bilayers is expected to affect the interactions of the peptides with the membrane because the dye molecules bear a net negative charge,(19) which should interact electrostatically with Arg9 and Lys9. This idea was explored by varying the concentration of dye in the bilayer (Figure S4). It was found that increasing the concentration of dye decreased the measured dissociation constant. Extrapolating the KD value to conditions without any dye in the membrane yielded a KD value of 60 μM. Control experiments were performed in the absence of membrane-bound dye by using total internal reflection fluorescence microscopy (TIRFM) with TAMARA-labeled Lys9 and Arg9. These results, which are discussed in the Supporting Information, yielded KD = 70 μM for Arg9, while also showing that Lys9 did not interact with the membrane (Figure S5). Such data is in very good agreement with the pH modulation data. To help understand the origin of the different binding behaviors of Lys9 and Arg9, molecular dynamics simulations were performed.
The results for simulations of POPC bilayers in the presence of both Arg9 and Lys9 are provided in Figure 5. The y-axis in each plot shows the number density profiles for selected chemical groups in zwitterionic POPC bilayers in the presence of six peptides (Figure 5a,c) and 12 peptides (Figure 5b,d), respectively, for the Arg9 and Lys9/128 lipid systems as a function of the distance from the bilayer center. In the case of arginine-rich peptides, an increased number density occurred in the bilayer headgroup region where the choline and phosphate groups are located (Figure 5). Increasing the peptide concentration increased the peptide number density in the bilayer headgroup region (compare red curves in the number density profiles in Figure 5a,c), indicative of increased adsorption of Arg9 at higher peptide concentrations. It should be noted that the peptide concentration at the membrane surface was only slightly enhanced compared to its number density in the bulk region away from the bilayer. In the case of Lys9, the MD simulations demonstrated that the peptides did not adsorb at all (red lines in the number density profiles in Figure 5b,d), which is in agreement with the experimental results (Figure 4). In fact for Lys9, the peptide number density at the membrane surface was depleted compared to that in the bulk region, which indicates that the free energy of interaction of lysine-rich peptides at zwitterionic POPC bilayers is unfavorable. The adsorption of arginine-rich peptides compared with lysine-rich peptides suggests that the interactions of the guanidinium moieties on the arginine side chain with the zwitterionic bilayer were both ion-specific and decisive. In fact, it appears that the ability of the guanidinium moiety on Arg9 to interact in a bidentate fashion with the phosphate moiety on the lipid headgroup is extremely important compared to the interactions of the amino groups of Lys9, which can only interact with one headgroup at a time. These MD results obtained with the AMBER99SB + SLIPIDS force field are consistent with the experimental data in Table 1, which is not the case for previous MD simulations with the OPLS/AA + Berger force field where it was predicted that lysine-rich peptides should also weakly adsorb to neutral zwitterionic POPC bilayers.(37)
Figure 5.
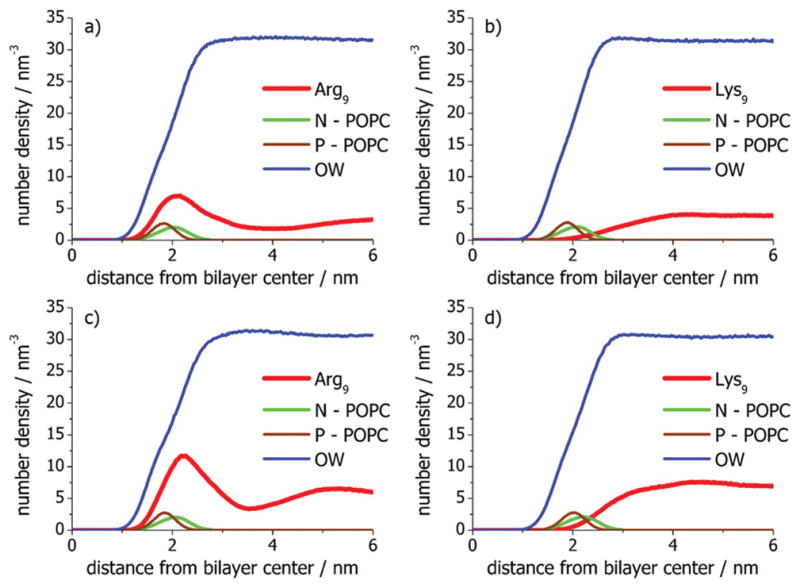
Number density profiles for Arg9 and Lys9, POPC choline nitrogen atom (N), POPC phosphate P atom (P), and the oxygen water atom (OW) in pure POPC bilayers as a function of the distance from the bilayer center. Simulations were performed at low peptide concentration with both (a) Arg9 and (b) Lys9. Additional simulations with (c) Arg9 and (d) Lys9 were also run with twice the amount of peptide.
Table 1.
Apparent Dissociation Constants KD, Hill Coefficients of Cooperativity n, and Apparent pKa Values for POPC Lipid Bilayers with Varying Concentrations of POPG Lipids
| mol % POPG + mol% ortho-rhodamine B | Lys9 | Arg9 | ortho-rhodamine B apparent pKa | ||
|---|---|---|---|---|---|
| KD(μM) | n | KD(μM) | n | ||
| 0 + 0 | NA | NA | 70 ± 19a,b | 0.75a,b | NA |
| 0 + 0.5 | NA | NA | 0.89 ± 0.38a | 0.39a | 6.2 |
| 5 + 0.5 | 77 ± 20a | 0.12a | 0.46 ± 0.047a | 0.68a | 6.3 |
| 10 + 0.5 | 55 ± 3 | 0.22 | 0.12 ± 0.016 | 0.73 | 6.6 |
| 20 + 0.5 | 2.5 ± 0.5 | 0.24 | 0.039 ± 0.005 | 1.0 | 6.9 |
| 30 + 0.5 | 2.1 ± 0.8 | 0.19 | 0.029 ± 0.007 | 1.0 | 7.1 |
Assay performed at pH 6.4; all others performed at pH 6.8.
Assay performed with TAMRA dye labeling, rather than membrane labeling.
Arg9 and Lys9 Binding Studies with POPG and POPC
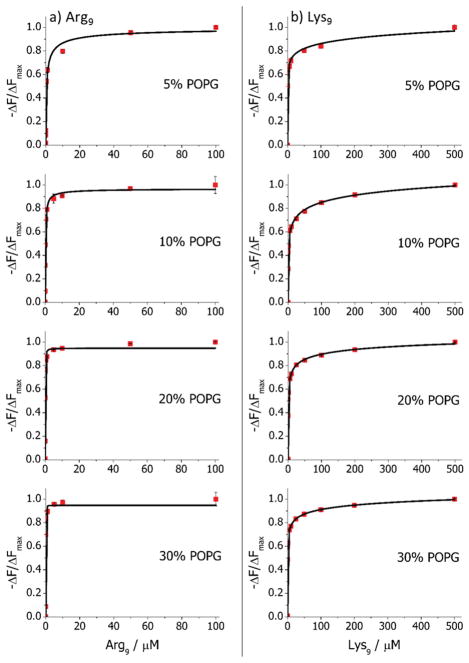
Next, experiments were performed to probe the interactions of Arg9 and Lys9 with mixed POPG/POPC bilayers in a manner analogous to the procedure described above with pure POPC (Figure 6). Specifically, experiments were conducted with POPG concentrations ranging from 10 to 30 mol % in 10 mM PBS at pH 6.8 with 150 mM NaCl in the same type of flow cell devices. Experiments were also performed at pH 6.4 with 5.0 mol % POPG. It should be noted that the slightly higher pH employed with higher concentrations of POPG was necessary to keep the assay within the linear sensing range of the ortho-rhodamine B probe (see Figure S2 for details). For Arg9, the apparent KD values extracted from this isotherm decreased from 460 to 29 nM as the concentration of POPG in the membrane was increased from 5 to 30 mol % (Figure 6a and Table 1). Moreover, both the 5 and 10 mol % POPG bilayers gave fluorescence response signals that were best fits to Hill–Waud isotherms showing modest negative cooperativity. Most strikingly, the interactions of Arg9 with bilayers composed of 20 and 30 mol % POPG were best fit with Hill coefficients of 1.0, demonstrating no anticooperative effect but instead simple Langmuir isotherm behavior.
Figure 6.
Binding isotherms for (a) Arg9 and (b) Lys9 interactions with bilayers containing 0.5 mol % ortho-rhodamine B POPE in POPC with varying concentrations of POPG. Experiments were performed in 10 mM PBS with 150 mM NaCl at pH 6.8 for 10, 20, and 30 mol % POPG and at pH 6.4 for 5 mol % POPG.
For Lys9, the apparent KD values extracted from these isotherms decreased from 77 to 2.1 μM as the concentration of POPG in the membrane was increased from 5 to 30 mol % (Figure 6b and Table 1). Moreover, the n values in all cases were well below unity, indicating that the binding process was strongly anticooperative. This finding is consistent with previous results where it has been reported that vesicles containing anionic lipids can reverse their surface potential (from negative to positive) upon incubation with a sufficient concentration of cationic peptides.(38, 39)Therefore, as more peptides bind, they experience ever greater electrostatic repulsion from the surface as depicted schematically in Figure 1b. Also, it is known that Lys9 will not penetrate beyond the membrane headgroup region.(40–42) It may be the case that the inability of the peptide to penetrate the bilayers is an additional factor in the anticooperative nature of the binding.
As noted above, Arg9 and Lys9 experiments involving 5 mol % POPG were performed at pH 6.4 instead of 6.8. As seen from Table 1, the Hill coefficient in this case, especially for Lys9, is somewhat lower than those for the other three POPG concentrations. Increasing the charge on the peptide by lowering the pH should increase the binding affinity to anionic bilayers for the initial peptides that adsorb but also will lead to greater anticooperativity as subsequent peptides absorb and attenuate the surface charge. This is likely the cause of the modestly greater anticooperativity which is observed. It is known that decreasing the ionic strength of the solution results in tighter binding for these systems, which would also speak to an electrostatic effect.(38) Additionally, a lower mol % of POPG in the bilayer should affect the average number of bound lipids per peptide,(43) which may be indicated by a lower Hill coefficient in these experiments.
Comparison of the apparent KD values for Arg9 with Lys9 reveals significantly tighter affinity for the former with bilayers containing POPG (Table 1). In fact, the equilibrium dissociation constants are roughly 2 orders of magnitude lower for Arg9 compared with Lys9, although these differences were greater at 5 and 10 mol % POPG compared with 20 and 30 mol %. Additionally, the values of n for Arg9 associating with 5 and 10 mol % POPG were much closer to unity compared with the corresponding cases with Lys9. The most striking difference, however, is at 20 and 30 mol % POPG, where the Arg9 binding becomes noncooperative, while the Lys9 binding remains strongly anticooperative. MD simulations were performed to help understand these phenomena, as described below.
MD simulations were run for both Arg9 and Lys9 with bilayers containing 30 mol % POPG at two different peptide concentrations (i.e., with 6 and 12 peptides). For Arg9, the partitioning of the peptide to the lipid/aqueous interface was quite pronounced and became even higher as the peptide concentration was increased (Figure 7a,c). The adsorption of Lys9 was also enhanced at the membrane surface (Figure 7b,d), but the effect was weaker than the adsorption of arginine-rich peptides as seen in the peptide number density profiles. These results, again, clearly demonstrate the role of ion specific interactions in addition to electrostatic forces, which certainly are also crucial in the absorption of the peptides at the interface.
Figure 7.
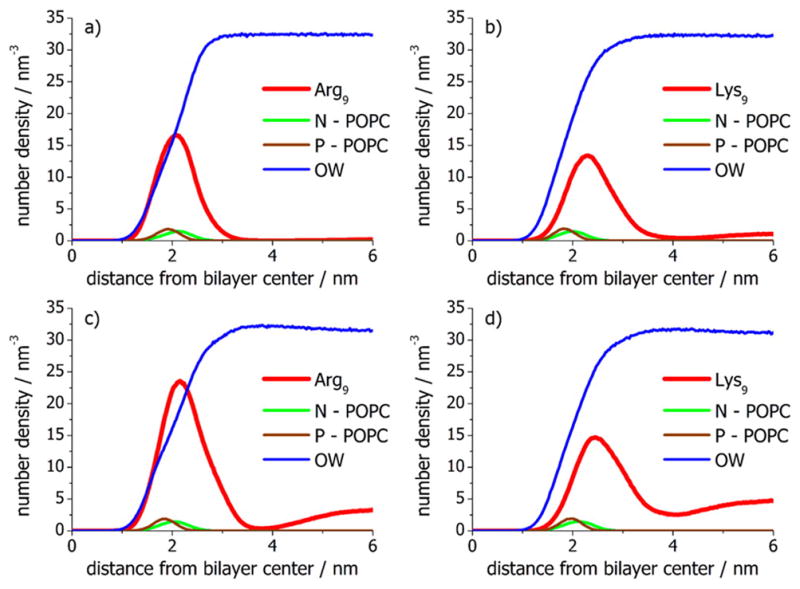
Number density profiles for Arg9 and Lys9, POPC choline nitrogen atom, POPC phosphate P atom (P), and oxygen water atom (OW) in 70 mol % POPC + 30 mol % POPG bilayers as a function of the distance from the bilayer center. Simulations were performed at low peptide concentration with both (a) Arg9 and (b) Lys9. Additional simulations with (c) Arg9 and (d) Lys9 were also run with twice the amount of peptide.
Examination of the MD number density profiles above shows good qualitative agreement with the experimental data presented in the Table 1. In particular, the peptide number density profiles suggest that there is an increased propensity of Arg9 to partition to the interface compared to Lys9for bilayers containing POPG. On the basis of the MD simulations, this enhanced absorption of arginine over lysine at the bilayer surface should be directly related to the ability of arginine-rich peptides to bind more favorably to bilayers due to interactions of the guanidinium moiety with the polar groups in the bilayer in contrast to lysine-rich peptides.(37) Moreover, more favorable guanidinium–guanidinium interactions compared with amino group–amino group interactions represent an important factor which contributes to the enhanced adsorption of Arg9.(37) These points are explored in more detail in the Discussion section.
Discussion
The structural makeup of the Lys9 and Arg9 peptides is similar in many ways. Both peptides are highly positively charged and well-known to associate with anionic phospholipids, especially phosphatidylserine and PG.(38) These two peptides, however, show markedly different behavior, both in the strength and mode of their interactions with membranes. It has been shown that although found in cell-penetrating peptide sequences such as the TAT domain of HIV, lysine does not display a propensity to translocate across membranes to become internalized as a homopolymer (polylysine).(44) By contrast, arginine has been found not only to improve the membrane-penetrating capacity of polypeptide sequences such as the TAT domain when it is substituted in place of the lysine residues(45) but also to enable membrane-translocation behavior as a homopolymer (polyarginine) as well.(46) The difference in behavior between these two amino acids clearly lies in the different chemical properties of their cationic moieties.
The binding of arginine-rich peptides to negatively charged lipid membranes is more complex than the binding of polylysine. This can be attributed primarily to the chemical specificity of the guanidinium moieties on its side chains.(47) Moreover, Arg–Arg interactions can overcome like-charge repulsion via their capacity for like-charge ion pairing, as found in some protein structures.(17, 48) In fact, guanidinium ions have been specifically shown to form parallel stacks in solution,(49) and both molecular dynamic simulations and ab initio calculations have confirmed the ability of guanidinium ion pairing in arginine-rich peptides as well as in solution.(15, 16) Guanidinium, being a planar ion, forms in-plane hydrogen bonds upon binding to anionic membranes, and parallel stacking can lead to like-charge association between guanidinium ions via the weakly hydrated face of the ion.(50) Calorimetric data has suggested that the association of polyarginine peptides with PG lipids may involve hydrophobic interactions,(51) and it follows that with increasing concentrations of PG in the bilayer one would expect close range interactions to rise in significance relative to long-range electrostatic repulsions. The data in Figures 6 and 7 as well as Table 1 are consistent with the notion that anticooperative effects due to electrostatic repulsion are, in fact, overcome at sufficiently high POPG concentrations where the short-range ion-pairing interactions of arginine are sufficiently attractive to just balance electrostatic repulsion. In other words, it appears that the generic electrostatic like-charge repulsions are nearly exactly offset by specific guanidinium–guanidinium ion pairing effects to yield peptide–membrane interactions that display noncooperative Langmuir isotherm behavior when sufficient POPG is present.
In theory, it is possible to account for the electrostatic repulsion of peptides from the bilayer surface by plotting the change in fluorescence as a function of the peptide concentration in the double layer above the bilayer interface. The interfacial peptide concentration can be determined using Gouy–Chapman theory; however, this requires the surface charge of the bilayer to be known at every point on the titration curve. While our label-free assay can detect binding events, it is currently unable to quantify the absolute surface density of peptides at the bilayer interface. This makes determining the surface charge on the bilayer, and therefore the interfacial peptide concentration, quite difficult. While plotting the change in fluorescence as a function of the interfacial peptide concentration would remove long-range electrostatic effects from the binding curves, this correction would not change the interpretation of our results. An electrostatic correction will cause the n values given by the Hill–Waud fits to the data for both Arg9 and Lys9 to increase, but the measured n values for Arg9 will increase as a function of PG content in the membrane while those for Lys9 should remain constant.
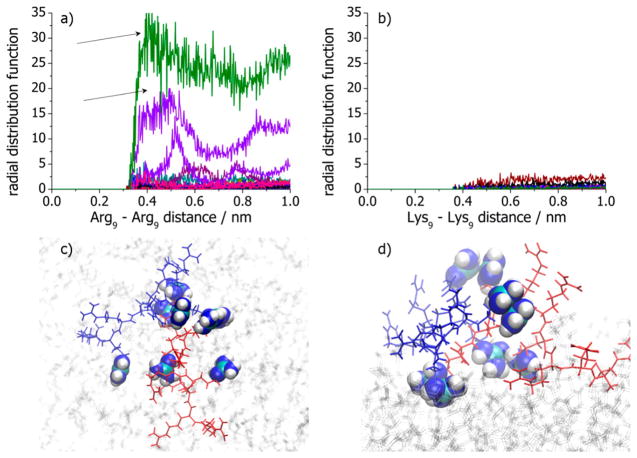
Additional information about ion pairing between Arg9 peptides on POPG-containing bilayers can be obtained by plotting the radial distribution function of the central carbon atoms for pairs of guanidinium moieties on separate peptides. Such functions for selected pairs are shown in Figure 8a. As can be seen, a maximum tends to occur near 0.4–0.5 nm separation, which resembles the like-charge pairing of guanidinium groups.(15, 16, 37) This is consistent with aggregation and dimer formation of Arg9 on bilayer surfaces with 30 mol % POPG. In the case of lysine-rich peptides, there is no evidence for intermolecular interactions between the peptides or signatures for ion pairing between the amine groups (Figure 8b).(15, 16) Figure 8c shows a typical snapshot of an arginine dimer adsorbed onto a POPG-containing bilayer in a top-down view, while Figure 8d shows a side-on view for the same snapshot highlighting the guanidinium pair. The existence of arginine aggregates has also been suggested to occur in bulk solutions(37) but with shorter MD simulations which employed a Berger united atom force field for the lipids.
Figure 8.
(a) Radial distribution functions (RDFs) between central carbon atoms in different Arg9 peptides in the 12 peptides/128 lipids Arg9–POPC+POPG system. The RDFs marked by arrows denote direct contact pairs between central carbon atoms in guanidinium moieties present in long-living Arg9–Arg9 aggregates. (b) Same plot as panel a for the central nitrogen atoms on different Lys9 peptides. Snapshots of (c) top-down view and (d) side-on view of two individual peptides (depicted in the blue and red stick drawings) is shown forming an aggregate in the 12 peptides/128 lipids Arg9–POPC+POPG system. The guanidinium groups in close contact are shown with a van der Waals representation.
The charged amino group on lysine can be expected to interact with lipid interfaces predominately via electrostatic forces, including hydrogen bonding, although there is evidence for some hydrophobic interactions as well.(10) The thermodynamic data shown herein, however, clearly demonstrate that this peptide experiences strongly anticooperative binding to bilayers containing a wide range of POPG concentrations. Such behavior is consistent with the known gradual reversal of the interfacial potential of the membrane as more peptide is introduced.(39) Indeed, as Lys9residues begin to populate the surface, each peptide is predicted to bind up to eight PG lipids.(38)Therefore, once all the surface POPG moieties are occupied, a reversal of the surface potential may occur. In fact, a reversal of the charge on POPG vesicles has been previously demonstrated with Lys5.(38)
Conclusions
Herein the interactions of Arg9 and Lys9 were explored with zwitterionic (POPC) and negatively charged mixed POPC/POPG lipid bilayers. The studies were conducted using fluorescence binding assays combined with all-atom molecular dynamics simulations. The experimentally determined apparent dissociation constants and Hill cooperativity coefficients clearly showed that Arg9 exhibited only weakly anticooperative binding at the bilayer interface at lower POPG concentrations, with the anticooperativity vanishing when 20 mol % POPG was present. By contrast, Lys9 exhibited strongly anticooperative binding behavior at all POPG concentrations. Moreover, the dissociation constants of Lys9 were found to be approximately 2 orders of magnitude lower than those for Arg9. The stronger and more cooperative interactions for Arg9appeared to occur both because of stronger guanidinium interactions with the lipid head groups as well as the like ion pairing ability for the guanidinium moieties.
The results found for Arg9 and Lys9 interactions with POPG-containing membranes may have wider biophysical implications. For example, the ability of guanidinium moieties on arginine to interact favorably at higher concentrations on the bilayer surface may provide clues to the mechanism for entry of cell-penetrating peptides into cells. One hypothesis is that Arg-containing peptides can form transient toroidal pores in the membrane surface.(52, 53) Such a mechanism is consistent with the stacking interactions found herein whereby electrostatic repulsions were overcome at shorter distances. By contrast, Lys was not able to form these types of stacking interactions, and pore formation and peptide entry would not be able to occur via this pathway. Other hypotheses can also be tested using simple model studies like the ones described herein. Understanding the mechanisms for membrane penetration in synthetic peptides as well as in naturally occurring proteins such as penetratin or the Tat protein remains an important challenge with implications for the ability to efficiently deliver drugs across cell membranes.(4) Also, the ability to elucidate interaction specifics such as the hydrophobic stacking ability of arginine will allow for a more detailed understanding of these systems and should aid in the design of more effective molecules for use in CPP strategies.
Supplementary Material
Acknowledgments
P.S.C. thanks the Office of Naval Research (N00014-14-1-0792) for funding. P.J. thanks the Czech Science Foundation (grant no. 16-01074S) and the Academy of Sciences of the Czech Republic for the Praemium Academie award. P.J. and M.V. also acknowledge the Academy of Finland for the FiDiPro award. M.V. is grateful to the Croatian Academy of Science and Arts for the financial support and CSC–IT Centre for Science (Espoo, Finland) for computational resources (project no. tty2030).
References
This article references 53 other publications.
- 1.Yount NY, Yeaman MR. Multidimensional Signatures in Antimicrobial Peptides. Proc Natl Acad Sci U S A. 2004;101:7363–7368. doi: 10.1073/pnas.0401567101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blondelle SE, Lohner K, Aguilar MI. Lipid-Induced Conformation and Lipid-Binding Properties of Cytolytic and Antimicrobial Peptides: Determination and Biological Specificity. Biochim Biophys Acta, Biomembr. 1999;1462:89–108. doi: 10.1016/S0005-2736(99)00202-3. [DOI] [PubMed] [Google Scholar]
- 3.Epand RM, Vogel HJ. Diversity of Antimicrobial Peptides and Their Mechanisms of Action. Biochim Biophys Acta, Biomembr. 1999;1462:11–28. doi: 10.1016/S0005-2736(99)00198-4. [DOI] [PubMed] [Google Scholar]
- 4.Koren E, Torchilin VP. Cell-Penetrating Peptides: Breaking through to the Other Side. Trends Mol Med. 2012;18:385–393. doi: 10.1016/j.molmed.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Rapaport D, Shai Y. Interaction of Fluorescently Labeled Pardaxin and Its Analogs with Lipid Bilayers. J Biol Chem. 1991;266:23769–23775. [PubMed] [Google Scholar]
- 6.Ludtke SJ, He K, Heller WT, Harroun TA, Yang L, Huang HW. Membrane Pores Induced by Magainin. Biochemistry. 1996;35:13723–13728. doi: 10.1021/bi9620621. [DOI] [PubMed] [Google Scholar]
- 7.Gazit E, Miller IR, Biggin PC, Sansom MSP, Shai Y. Structure and Orientation of the Mammalian Antibacterial Peptide Cecropin P1 within Phospholipid Membranes. J Mol Biol. 1996;258:860–870. doi: 10.1006/jmbi.1996.0293. [DOI] [PubMed] [Google Scholar]
- 8.Bechinger B, Lohner K. Detergent-Like Actions of Linear Amphipathic Cationic Antimicrobial Peptides. Biochim Biophys Acta, Biomembr. 2006;1758:1529–1539. doi: 10.1016/j.bbamem.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 9.Bechara C, Sagan S. Cell-Penetrating Peptides: 20 Years Later, Where Do We Stand? FEBS Lett. 2013;587:1693–1702. doi: 10.1016/j.febslet.2013.04.031. [DOI] [PubMed] [Google Scholar]
- 10.Hoernke M, Schwieger C, Kerth A, Blume A. Binding of Cationic Pentapeptides with Modified Side Chain Lengths to Negatively Charged Lipid Membranes: Complex Interplay of Electrostatic and Hydrophobic Interactions. Biochim Biophys Acta, Biomembr. 2012;1818:1663–1672. doi: 10.1016/j.bbamem.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 11.Rothbard JB, Jessop TC, Lewis RS, Murray BA, Wender PA. Role of Membrane Potential and Hydrogen Bonding in the Mechanism of Translocation of Guanidinium-Rich Peptides into Cells. J Am Chem Soc. 2004;126:9506–9507. doi: 10.1021/ja0482536. [DOI] [PubMed] [Google Scholar]
- 12.Sakai N, Matile S. Anion-Mediated Transfer of Polyarginine across Liquid and Bilayer Membranes. J Am Chem Soc. 2003;125:14348–14356. doi: 10.1021/ja037601l. [DOI] [PubMed] [Google Scholar]
- 13.Jung H, Robison AD, Cremer PS. Detecting Protein-Ligand Binding on Supported Bilayers by Local Ph Modulation. J Am Chem Soc. 2009;131:1006–1014. doi: 10.1021/ja804542p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Masunov A, Lazaridis T. Potentials of Mean Force between Ionizable Amino Acid Side Chains in Water. J Am Chem Soc. 2003;125:1722–1730. doi: 10.1021/ja025521w. [DOI] [PubMed] [Google Scholar]
- 15.Vazdar M, Vymetal J, Heyda J, Vondrasek J, Jungwirth P. Like-Charge Guanidinium Pairing from Molecular Dynamics and Ab Initio Calculations. J Phys Chem A. 2011;115:11193–11201. doi: 10.1021/jp203519p. [DOI] [PubMed] [Google Scholar]
- 16.Vazdar M, Uhlig F, Jungwirth P. Like-Charge Ion Pairing in Water: An Ab Initio Molecular Dynamics Study of Aqueous Guanidinium Cations. J Phys Chem Lett. 2012;3:2021–2024. doi: 10.1021/jz3007657. [DOI] [Google Scholar]
- 17.Vondrasek J, Mason PE, Heyda J, Collins KD, Jungwirth P. The Molecular Origin of Like-Charge Arginine-Arginine Pairing in Water. J Phys Chem B. 2009;113:9041–9045. doi: 10.1021/jp902377q. [DOI] [PubMed] [Google Scholar]
- 18.Xia YN, McClelland JJ, Gupta R, Qin D, Zhao XM, Sohn LL, Celotta RJ, Whitesides GM. Replica Molding Using Polymeric Materials: A Practical Step toward Nanomanufacturing. Adv Mater. 1997;9:147–149. doi: 10.1002/adma.19970090211. [DOI] [Google Scholar]
- 19.Xia YN, Whitesides GM. Soft Lithography. Angew Chem, Int Ed. 1998;37:550–575. doi: 10.1002/(SICI)1521-3773(19980316)37:5<550::AID-ANIE550>3.3.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 20.Huang D, Zhao T, Xu W, Yang T, Cremer PS. Sensing Small Molecule Interactions with Lipid Membranes by Local Ph Modulation. Anal Chem. 2013;85:10240–10248. doi: 10.1021/ac401955t. [DOI] [PubMed] [Google Scholar]
- 21.Axelrod D, Burghardt TP, Thompson NL. Total Internal Reflection Fluorescence. Annu Rev Biophys Bioeng. 1984;13:247–268. doi: 10.1146/annurev.bb.13.060184.001335. [DOI] [PubMed] [Google Scholar]
- 22.Yang TL, Jung SY, Mao HB, Cremer PS. Fabrication of Phospholipid Bilayer-Coated Microchannels for on-Chip Immunoassays. Anal Chem. 2001;73:165–169. doi: 10.1021/ac000997o. [DOI] [PubMed] [Google Scholar]
- 23.Burmeister JS, Olivier LA, Reichert WM, Truskey GA. Application of Total Internal Reflection Fluorescence Microscopy to Study Cell Adhesion to Biomaterials. Biomaterials. 1998;19:307–325. doi: 10.1016/S0142-9612(97)00109-9. [DOI] [PubMed] [Google Scholar]
- 24.Hlady V, Reinecke DR, Andrade JD. Fluorescence of Adsorbed Protein Layers: I. Quantitation of Total Internal Reflection Fluorescence. J Colloid Interface Sci. 1986;111:555–569. doi: 10.1016/0021-9797(86)90059-7. [DOI] [Google Scholar]
- 25.Mark JE. Polymer Data Handbook. 2. Oxford University Press; Oxford, U.K: 2009. [Google Scholar]
- 26.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of Simple Potential Functions for Simulating Liquid Water. J Chem Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 27.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of Multiple Amber Force Fields and Development of Improved Protein Backbone Parameters. Proteins: Struct, Funct, Genet. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jämbeck JPM, Lyubartsev AP. Derivation and Systematic Validation of a Refined All-Atom Force Field for Phosphatidylcholine Lipids. J Phys Chem B. 2012;116:3164–3179. doi: 10.1021/jp212503e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jämbeck JPM, Lyubartsev AP. An Extension and Further Validation of an All-Atomistic Force Field for Biological Membranes. J Chem Theory Comput. 2012;8:2938–2948. doi: 10.1021/ct300342n. [DOI] [PubMed] [Google Scholar]
- 30.Jämbeck JPM, Lyubartsev AP. Another Piece of the Membrane Puzzle: Extending Slipids Further. J Chem Theory Comput. 2013;9:774–784. doi: 10.1021/ct300777p. [DOI] [PubMed] [Google Scholar]
- 31.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A Smooth Particle Mesh Ewald Method. J Chem Phys. 1995;103:8577–8593. doi: 10.1063/1.470117. [DOI] [Google Scholar]
- 32.Parrinello M, Rahman A. Polymorphic Transitions in Single Crystals: A New Molecular Dynamics Method. J Appl Phys. 1981;52:7182–7190. doi: 10.1063/1.328693. [DOI] [Google Scholar]
- 33.Nose S. A Molecular Dynamics Method for Simulations in the Canonical Ensemble. Mol Phys. 1984;52:255–268. doi: 10.1080/00268978400101201. [DOI] [Google Scholar]
- 34.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A Linear Constraint Solver for Molecular Simulations. J Comput Chem. 1997;18:1463–1472. doi: 10.1002/(SICI)1096-987X(199709)18:12<1463::AID-JCC4>3.0.CO;2-H. [DOI] [Google Scholar]
- 35.Hockney RW, Goel SP, Eastwood J. Quiet High Resolution Computer Models of a Plasma. J Comput Phys. 1974;14:148–158. doi: 10.1016/0021-9991(74)90010-2. [DOI] [Google Scholar]
- 36.Hess B, Kutzner C, van der Spoel D, Lindahl E. Gromacs 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 37.Vazdar M, Wernersson E, Khabiri M, Cwiklik L, Jurkiewicz P, Hof M, Mann E, Kolusheva S, Jelinek R, Jungwirth P. Aggregation of Oligoarginines at Phospholipid Membranes: Molecular Dynamics Simulations, Time-Dependent Fluorescence Shift, and Biomimetic Colorimetric Assays. J Phys Chem B. 2013;117:11530–11540. doi: 10.1021/jp405451e. [DOI] [PubMed] [Google Scholar]
- 38.Kim JY, Mosior M, Chung LA, Wu H, Mclaughlin S. Binding of Peptides with Basic Residues to Membranes Containing Acidic Phospholipids. Biophys J. 1991;60:135–148. doi: 10.1016/S0006-3495(91)82037-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Murray D, Arbuzova A, Hangyas-Mihalyne G, Gambhir A, Ben-Tal N, Honig B, McLaughlin S. Electrostatic Properties of Membranes Containing Acidic Lipids and Adsorbed Basic Peptides: Theory and Experiment. Biophys J. 1999;77:3176–3188. doi: 10.1016/S0006-3495(99)77148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ben-Tal N, Honig B, Peitzsch RM, Denisov G, McLaughlin S. Binding of Small Basic Peptides to Membranes Containing Acidic Lipids: Theoretical Models and Experimental Results. Biophys J. 1996;71:561–575. doi: 10.1016/S0006-3495(96)79280-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roux M, Neumann JM, Bloom M, Devaux PF. 2H and 31P NMR Study of Pentalysine Interaction with Headgroup Deuterated Phosphatidylcholine and Phosphatidylserine. Eur Biophys J. 1988;16:267–273. doi: 10.1007/BF00254062. [DOI] [PubMed] [Google Scholar]
- 42.Bonev B, Watts A, Bokvist M, Grobner G. Electrostatic Peptide-Lipid Interactions of Amyloid-Beta Peptide and Pentalysine with Membrane Surfaces Monitored by P-31 Mas Nmr. Phys Chem Chem Phys. 2001;3:2904–2910. doi: 10.1039/b103352m. [DOI] [Google Scholar]
- 43.Mosior M, Mclaughlin S. Electrostatics and Reduction of Dimensionality Produce Apparent Cooperativity When Basic Peptides Bind to Acidic Lipids in Membranes. Biochim Biophys Acta, Biomembr. 1992;1105:185–187. doi: 10.1016/0005-2736(92)90178-O. [DOI] [PubMed] [Google Scholar]
- 44.Mitchell DJ, Kim DT, Steinman L, Fathman CG, Rothbard JB. Polyarginine Enters Cells More Efficiently Than Other Polycationic Homopolymers. J Pept Res. 2000;56:318–325. doi: 10.1034/j.1399-3011.2000.00723.x. [DOI] [PubMed] [Google Scholar]
- 45.Futaki S, Suzuki T, Ohashi W, Yagami T, Tanaka S, Ueda K, Sugiura Y. Arginine-Rich Peptides - an Abundant Source of Membrane-Permeable Peptides Having Potential as Carriers for Intracellular Protein Delivery. J Biol Chem. 2001;276:5836–5840. doi: 10.1074/jbc.M007540200. [DOI] [PubMed] [Google Scholar]
- 46.Wender PA, Mitchell DJ, Pattabiraman K, Pelkey ET, Steinman L, Rothbard JB. The Design, Synthesis, and Evaluation of Molecules That Enable or Enhance Cellular Uptake: Peptoid Molecular Transporters. Proc Natl Acad Sci U S A. 2000;97:13003–13008. doi: 10.1073/pnas.97.24.13003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang M, Waring AJ, Hong M. Phosphate-Mediated Arginine Insertion into Lipid Membranes and Pore Formation by a Cationic Membrane Peptide from Solid-State Nmr. J Am Chem Soc. 2007;129:11438–11446. doi: 10.1021/ja072511s. [DOI] [PubMed] [Google Scholar]
- 48.Magalhaes A, Maigret B, Hoflack J, Gomes JANF, Scheraga HA. Contribution of Unusual Arginine-Arginine Short-Range Interactions to Stabilization and Recognition in Proteins. J Protein Chem. 1994;13:195–215. doi: 10.1007/BF01891978. [DOI] [PubMed] [Google Scholar]
- 49.Mason PE, Neilson GW, Enderby JE, Saboungi ML, Dempsey CE, MacKerell AD, Brady JW. The Structure of Aqueous Guanidinium Chloride Solutions. J Am Chem Soc. 2004;126:11462–11470. doi: 10.1021/ja040034x. [DOI] [PubMed] [Google Scholar]
- 50.Mason PE, Brady JW, Neilson GW, Dempsey CE. The Interaction of Guanidinium Ions with a Model Peptide. Biophys J. 2007;93:L04–06. doi: 10.1529/biophysj.107.108290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwieger C, Blume A. Interaction of Poly(L-Arginine) with Negatively Charged Dppg Membranes: Calorimetric and Monolayer Studies. Biomacromolecules. 2009;10:2152–2161. doi: 10.1021/bm9003207. [DOI] [PubMed] [Google Scholar]
- 52.Herce HD, Garcia AE. Molecular Dynamics Simulations Suggest a Mechanism for Translocation of the Hiv-1 Tat Peptide across Lipid Membranes. Proc Natl Acad Sci U S A. 2007;104:20805–20810. doi: 10.1073/pnas.0706574105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Herce HD, Garcia AE, Litt J, Kane RS, Martin P, Enrique N, Rebolledo A, Milesi V. Arginine-Rich Peptides Destabilize the Plasma Membrane, Consistent with a Pore Formation Translocation Mechanism of Cell-Penetrating Peptides. Biophys J. 2009;97:1917–1925. doi: 10.1016/j.bpj.2009.05.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.