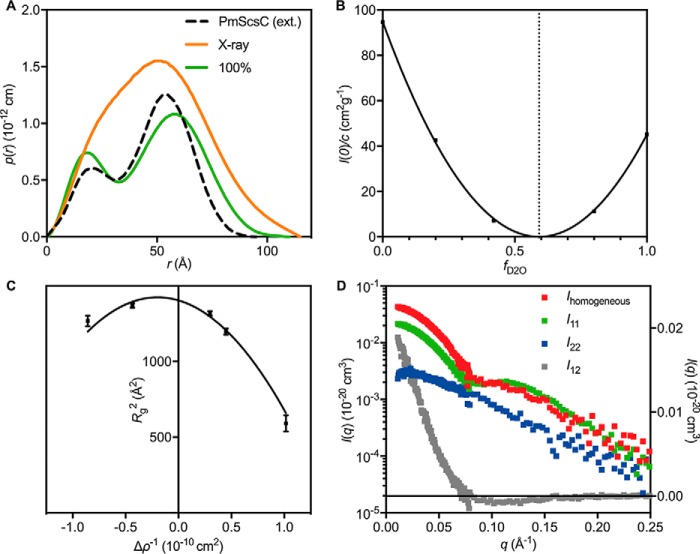
Figure 8.
Other SAXS/SANS results. A, p(r) derived from the experimental X-ray scattering data (orange) and the 100% scattering data (green), which should approximate the p(r) for PmScsC C87S alone. The p(r) derived from the extended (ext.) crystal structure (Protein Data Bank code 5ID4) is shown as a black dotted line for reference. B, a plot of I(0) normalized by concentration as a function of D2O content of the supporting solvent. The plot is parabolic in shape and reveals that the match point of the entire complex is 59% D2O (vertical dotted line). C, Stuhrmann plot for the 3:1 PmScsC C87S–DPmScsBα C114A complex, conforming to Rg2 = Rm2 + αΔρ−1 − βΔρ−2 where Rm is the radius of gyration of that object with homogenous contrast and α and β are related to the contrast variations within the object. The values obtained from a fit to the plot (solid black line) are: Rm = 37.5 ± 0.4, α = −210 ± 50, and β = 520 ± 100. The negative value of α reveals that the region with higher contrast (i.e. DPmScsBα C114A) lies toward the center of the molecule. Error bars represent the S.E. D, the composite scattering functions determined from the neutron contrast variation data. The I11 curve (green; left y axis) corresponds to scattering from PmScsC C87S, the I22 curve (blue; left y axis) corresponds to scattering from DPmScsBα C114A, and the I12 curve (gray; right y axis) is related to the arrangement of DPmScsBα C114A relative to PmScsC C87S. Ihomogeneous = I11 + I22 + I12 (red; left y axis) is the scattering curve of an object with the same shape as the PmScsC C87S–DPmScsBα C114A complex but with homogeneous contrast and is used for estimating the Porod volume and molecular mass from the SANS data.