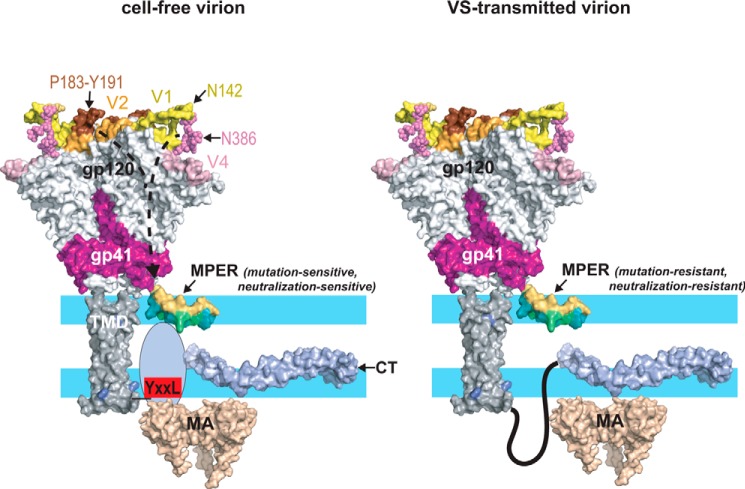
Figure 9.
Structural modulation of the MPER, a model. Left, depiction of the Env glycoprotein in a cell-free virion. The trimeric gp120–gp41 ectodomain was drawn using the coordinates from PDB code 5FYJ. The MPER (a monomer is shown for clarity) is embedded as a kinked helix in the polar headgroup (light blue)–acyl chain (white) interfacial region of the viral envelope as indicated by NMR studies of MPER peptides associated with membrane mimics (37, 38) and cryo-EM of HIV-1 particles (50). The hydrophobic acyl-chain–interactive face of the MPER helices is colored teal. The N-terminal segment of the CT (707–752) is unstructured in the absence of MA interactions (143) but becomes organized when engaged by MA trimers in hexameric arrays (62–64, 84). In this context, the MPER is in a conformation that is sensitive to mutations such as W666A, W672A, and I675A that potentially alter its interaction with the envelope. The effects of these mutations are mitigated by truncation of the CT or by Y712S, which uncouple Env from Gag due in part to disruption of the YXXL motif. In the SC54 isolate, the bridge formed between the V1-Asn142 and V4-Asn386 glycans and the V2–β-hairpin–stabilizing effects of the Pro183–Tyr191 interaction at the trimer apex confer a mutation-sensitive phenotype to the MPER via allosteric mechanisms. Right, in VS-transmitted virions, the MPER is resistant to mutations and neutralization by bNAb 10E8 perhaps due to an alternative CT conformation (108) and interaction with the MA domain. The various protein domains were drawn with PyMOL using the following coordinates: gp120–gp41 ectodomain, PDB code 5FYJ (120); MPER, PDB code 2PV6 (38); membrane-spanning sequence, PDB code 5JYN (144); CT, PDB code 5VWL (143); MA trimer, PDB code 1HIW (145).