Figure 4.
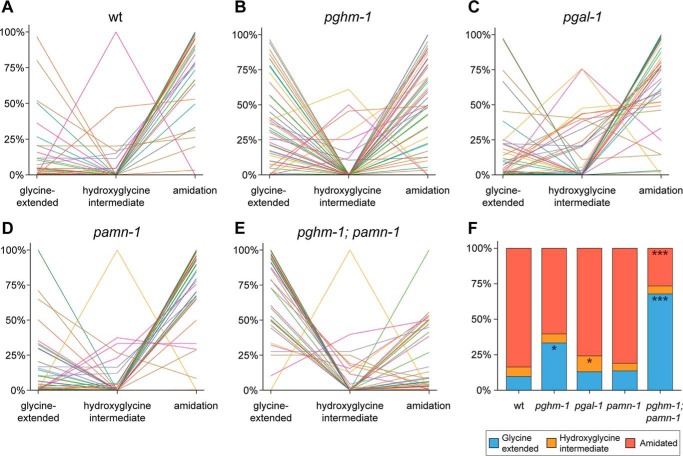
Modification states of separate identified neuropeptides (A–E) and distribution of neuropeptide modification states (F) in WT and mutant animals. Each individual line corresponds to an identified neuropeptide (52 in total). The y axis represents the mean values of normalized relative peptide abundances. The majority of neuropeptides in wildtype (wt) (A) are detected as carboxyl-terminal amidated peptides. Mutating pghm-1 seems to disrupt normal amidation, because more neuropeptides are detected with the carboxyl-terminal glycine still present (B). Knockout of pgal-1 seems to have only a small effect on the amount of amidated neuropeptides (C); however, an increase in hydroxyglycine intermediates can be seen. Inactivation of pamn-1 has no severe effects on amidation (D). Finally, mutating both pghm-1 and pamn-1 displays the most severe amidation effects (E). The majority of neuropeptides are found with their carboxyl-terminal glycine still present. When looking at the modification state distribution for all 52 peptides (F), WT animals indeed show a high occurrence of amidation (84.4%) and only low amounts of glycine-extended peptides. Although knockout of pghm-1 results in a rise of glycine-extended peptides (*, p < 0.05), other single mutants seem to be less affected. Although pgal-1 shows a significant increase in hydroxyglycine intermediates (*, p < 0.05), it still contains a high amount of amidated peptides. pamn-1 knockout does not seem to have any major effects on peptide amidation, because it closely resembles WT. A clear effect is seen in the pghm-1;pamn-1 double mutant, where the occurrence of amidated peptides collapses (***, p < 0.001) and is concomitant with a rise in glycine-extended peptides (***, p < 0.001). Because the effect in the double mutant is more severe than the additive effects of each single mutant, this may suggest that pghm-1 and pamn-1 are interchangeable to a certain degree. Data are derived from 52 identified peptides, present in all mutants. For each mutant, three (WT, pgal-1, and pamn-1) or two (pghm-1 and pghm-1;pamn-1) replicates were used. For more detail, including data of individual replicates and annotation of all individual neuropeptides, see Figs. S2 and S3.