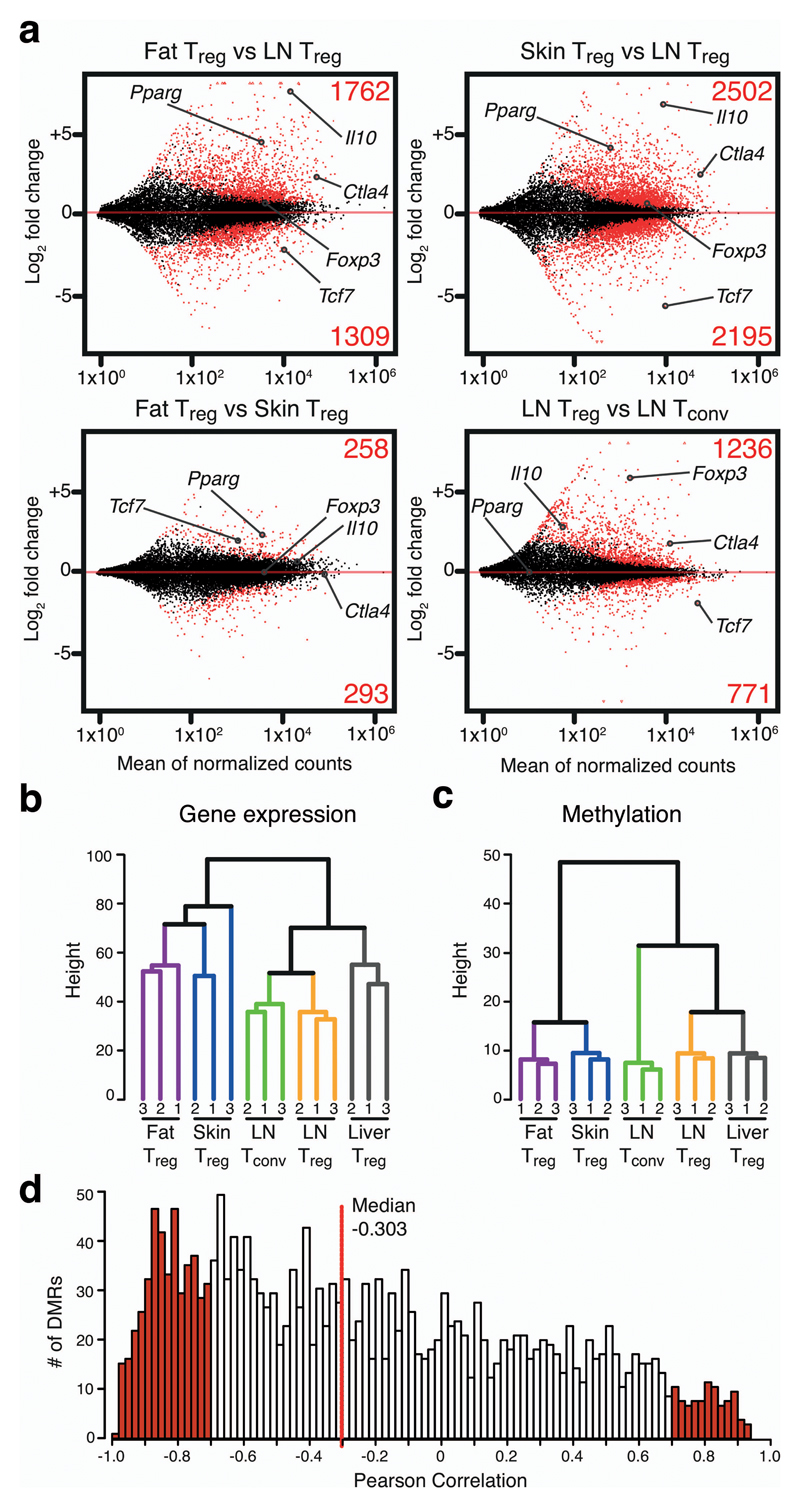
Figure 2. Transcriptome analysis of tissue-resident Treg cells and correlation with epigenetic dataset.
(a) MA plots calculated from RNA sequencing data of fat vs. LN Treg, skin Treg vs. LN Treg cells, fat vs. skin Treg, and LN Treg vs. Tconv cells. Numerators are plotted as mean of normalized counts. Fold changes are presented as log2. Significantly up- or downregulated genes (p<0.05) are highlighted in red, with the respective numbers shown above or below. Selected genes are labeled. (b) Unsupervised hierarchical clustering of gene expression data. Three replicates per group are shown, with numbers indicating the respective replicate. (c) Unsupervised hierarchical clustering of DMR methylation data. Three replicates per group are shown. (d) Pearson correlation between gene expression and DMR methylation. DMRs were intragenic or within 5kb of the nearest gene. Red line represents the median.