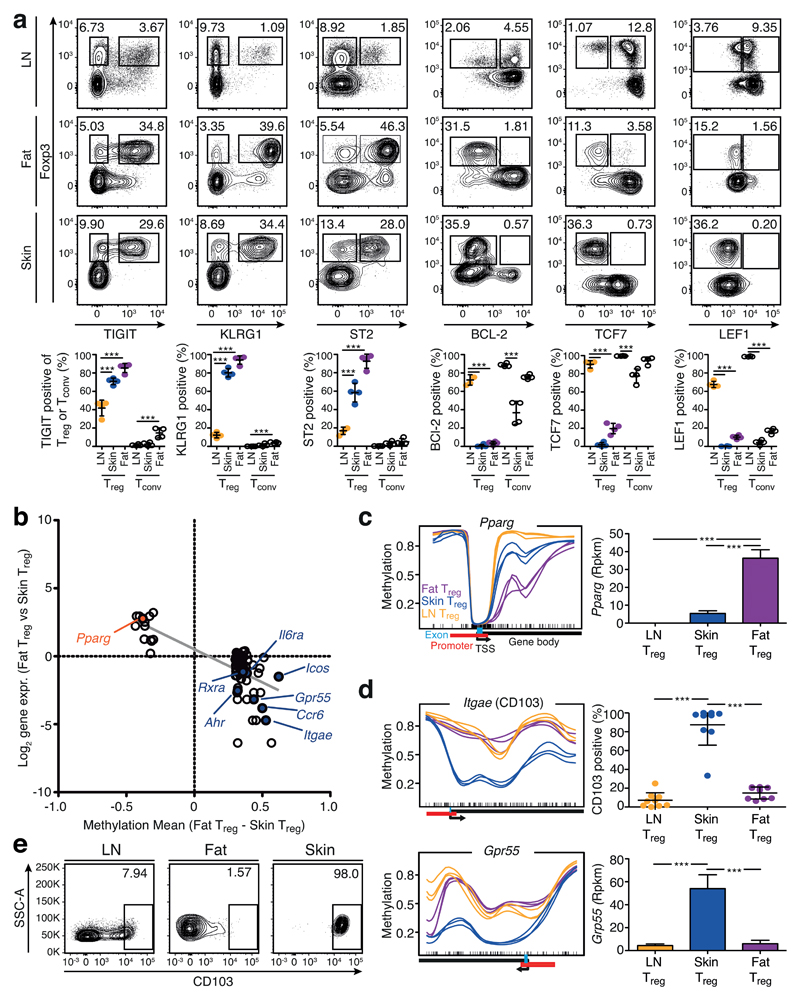
Figure 5. Validation of the common tissue Treg signature and identification of tissue-specific patterns.
(a) Flow cytometry analysis of TIGIT, KLRG1, ST2, BCL-2, TCF7 and LEF1 in fat, skin and LN Treg cells (CD19–MHCII–CD3+CD8–CD4+CD25+Foxp3+) and corresponding Tconv cells (CD19–MHCII–CD3+CD8–CD4+CD25– Foxp3–). Contour plots are concatenated files representative of four replicates, quantification at bottom part. Individual mice are shown (n=4). Statistical evaluation based on one-way ANOVA with Bonferoni post-test (***=p<0.001). (b) Differences in DMR methylation and gene expression of the comparison Treg cells from fat vs. skin as described in Fig. 3a. (c) Methylation pattern of the gene Pparg and corresponding gene expression. Shown are skin (blue), fat (purple), and LN Treg (yellow). Each line represents one individual replicate (n=3). Gene expression is plotted as reads per kilobase per million mapped reads (Rpkm). Significance based on RNA sequencing calculations as described in methods section and indicated by asterisks. (d) Methylation of Itgae (CD103) and Gpr55 in skin (blue), fat (purple), and LN Treg (yellow) as in (c). Quantification of CD103 data are based on (e), where contour plots indicate CD103 expression on Treg cells from LN, fat and skin measured via flow cytometry. Statistical evaluation based on one-way ANOVA with Bonferoni post-test (n=9). Means are shown with standard deviation (SD)