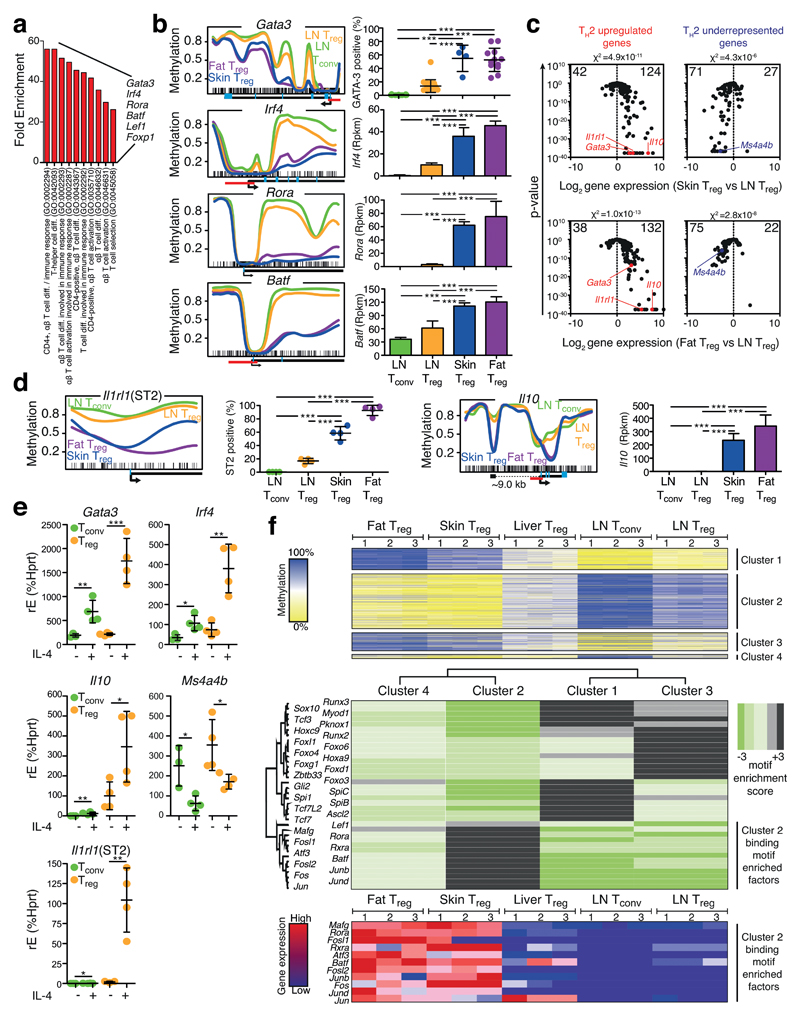
Figure 6. Fat and skin Treg cells are TH2-like polarized.
(a) Gene Ontology (GO) term analysis of the gene list described in Fig. 4b. (b) Methylation profile and protein (GATA-3) or gene expression for Gata3, Irf4, Rora and Batf as described in Fig. 3b. Average methylation values derived from three individual replicates are shown for fat Treg (purple), skin Treg (blue), LN Treg (yellow) and LN Tconv (green). (c) Diagnostic TH2 up- (red, left panel) or downregulated (blue, right panel) signature genes are plotted for the comparison skin vs. LN Treg (upper panel) and fat vs. LN Treg (lower panel). Numbers indicate the number of genes per site. Individual genes are highlighted. Chi2-test for proportions. (d) Methylation profile and protein or gene expression of Il1rl1 and Il10 as described in (b). ST2 expression quantification based on Fig. 5a and statistically evaluated with one-way ANOVA (n=4). (e) Culture of Treg or Tconv cells with 25 ng/mL IL-4 (+) or control (-) followed by qPCR-based evaluation of target gene expression. Statistical evaluation with unpaired two-tailed student’s t test (n=4). (f) Tissue Treg methylation data were grouped into 4 specific methylation-expression clusters (top panel). Based on these clusters, binding motif enriched factors were calculated and illustrated in a heat map (middle panel). Gene expression values of cluster 2 binding motif enriched factors for fat Treg, skin Treg, liver Treg, LN Tconv and LN Treg are shown as heatmap (bottom panel), where colors indicate relative expression (high=red; low=blue). Individual replicates are shown (n=3). Means are shown with SD.