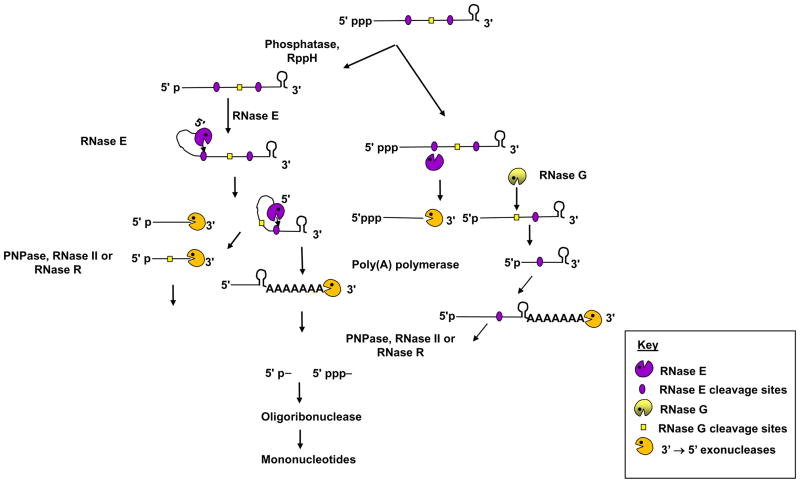
Fig. 2.
Model for the initiation of mRNA decay by RNase E. For the sake of simplicity, the other proteins associated with the RNase E-based degradosome are not shown. In addition, this model is independent of whether RNase E is associated with the inner membrane of E. coli. 5′ monophosphate RNA, a preferred substrate for RNase E is degraded via 5′ end dependent pathway. In contrast, 5′ triphosphate RNA is degraded via RNase E internal entry mechanism. Any endonucleolytically cleaved fragments with strong secondary structures, such as one containing a Rho-independent transcription terminator shown here, undergoes polyadenylation by PAP I. Subsequently, all decay intermediates are degraded by 3′ → 5′ exonucleases (PNPase, RNase II and RNase R) followed by oligoribonuclease to mononucleotides. Figure is not drawn to scale.