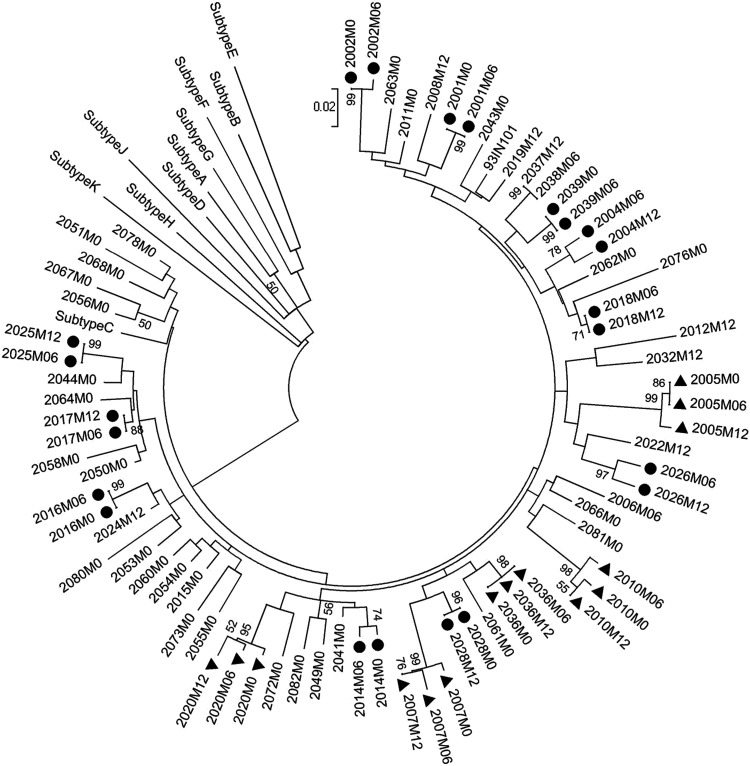
FIG. 2.
The phylogenetic analysis of the tat sequences. A total of 74 tat exon 1 sequences derived from 53 subjects are used in the analysis. Filled circles and filled triangles represent subjects from whom samples were available at two (n = 11) and three (n = 5) different time points, respectively. From all of the other subjects, samples at a single time point were collected. The reference sequences of tat from various genetic subtypes—A, B, C, D, E, F, G, H, J, K were downloaded from the Los Alamos HIV database. After gap stripping, there are a total of 201 nucleic acid positions in the final dataset. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method and are in the units of the number of base substitutions per site.