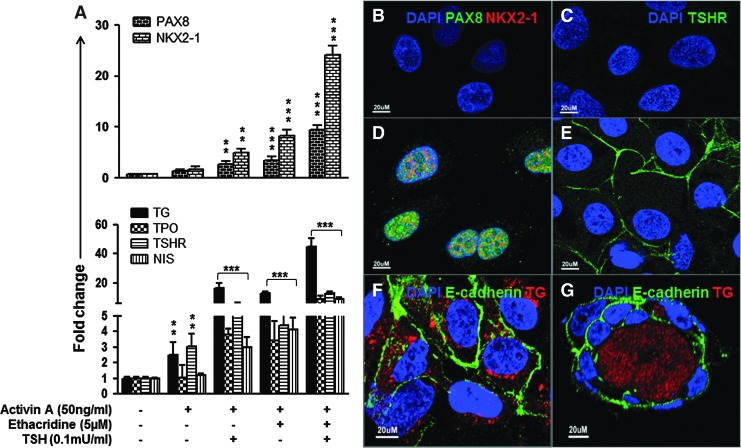
FIG. 4.
Analysis of differentiated hES cells. (A) qRT-PCR analysis for thyroid transcription factors (PAX8 and NKX2-1) and thyroid functional genes (TG, TPO, TSHR, and NIS) in differentiated cells. The relative expression of each transcript is presented as the fold change compared to undifferentiated ES cells (mean ± SEM) and represents one out of three separate experiments. Statistical analysis was performed by ANOVA, and the p-values were **p < 0.01 and ***p < 0.0001 for each gene when compared to untreated controls. (B–E) The immunostaining for (D) PAX8 (green) and NKX2-1(red) and (E) TSHR (green) in the differentiated hES cells and control cells (B and C). Note that the PAX8 and NKX2-1 expression is seen in the cell nuclei, and the TSHR expression is seen on the cell surface. Scale bar = 20 μm. (F) Immunodetection of TG (red) and E-cadherin (green) expression in differentiated hES cells. Note that the TG is seen in the cytoplasm, and E-cadherin is seen on the cell surface. Scale bar = 20 μm. (G) Immunodetection of TG (red) expression and E-cadherin (green) in a human thyroid neofollicle derived from differentiated hES cells. Note that the TG expression is seen in the intra-follicluar lumen, and E-cadherin expression is seen on the cell surface. Scale bar = 20 μm.