Figure 2. Ligand binding by a natural singlet PRPP riboswitch.
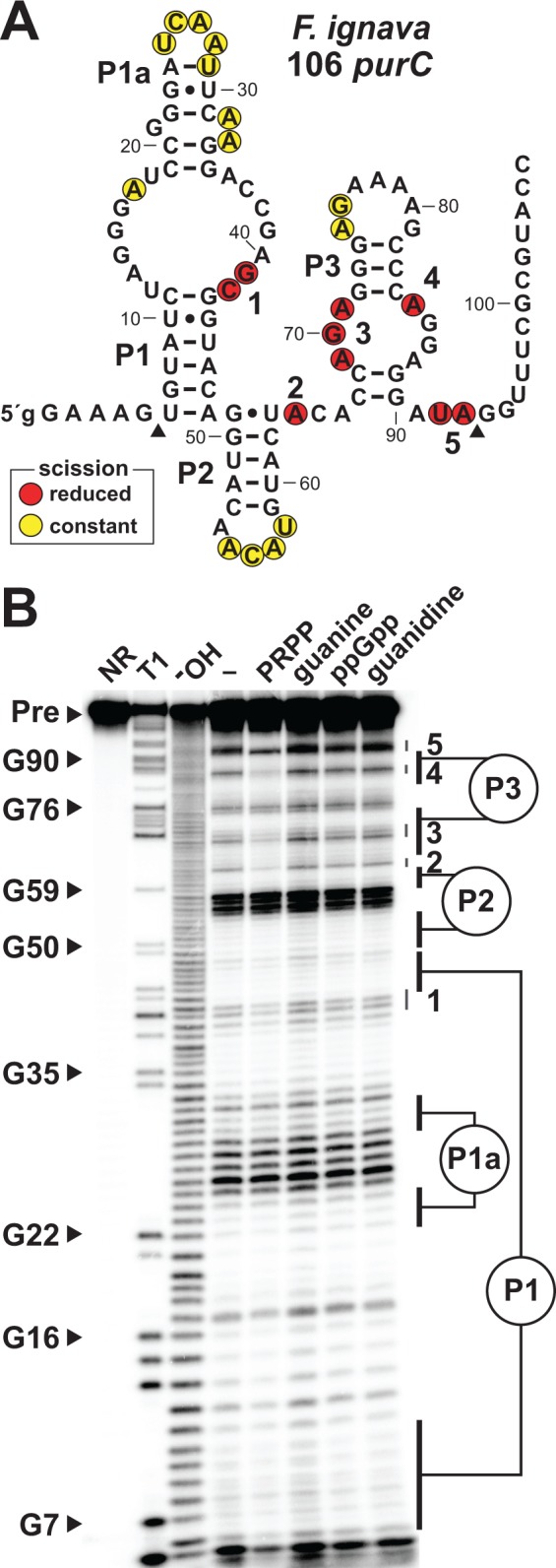
(A) Sequence and secondary structure of the 106 nucleotide RNA derived from the purC gene of F. ignava. Data collected in B were used to determine regions of constant or reduced scission upon the addition of PRPP to in-line probing assays. Lowercase g indicates a guanosine nucleotide added to enable efficient transcription by T7 RNA polymerase. Arrowheads demark the approximate range of nucleotides with in-line probing data available as depicted in B. (B) Polyacrylamide gel electrophoresis (PAGE) analysis of the spontaneous RNA cleavage products generated during in-line probing of 5′-32P-labeled 106 purC RNA. NR, T1 and –OH represent RNA undergoing no reaction, partial digest with RNase T1 (cleaves after guanosine nucleotides), or partial digest under alkaline conditions (cleaves after every nucleotide), respectively. Bands corresponding to selected G residues are annotated. In-line probing experiments contained either no ligand (–) or 1 mM of the compound indicated except guanine which was tested at 10 µM. Annotations 1 through 5identify prominent regions of the RNA that undergo structural stabilization in a PRPP-dependent manner. Data shown are representative of multiple experiments.
