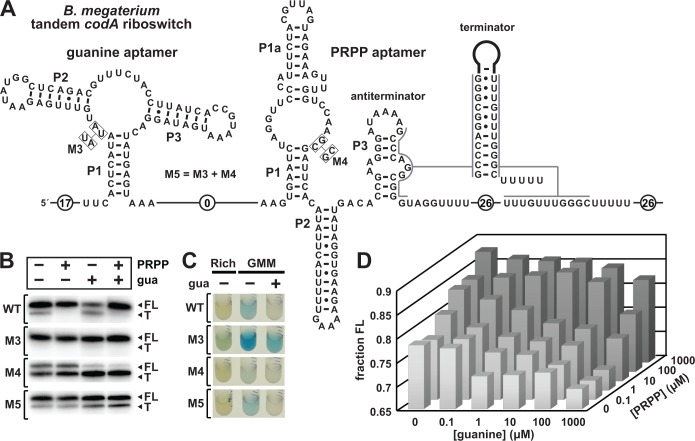
Figure 5. Guanine and PRPP competitively regulate transcript termination via a riboswitch integrating two aptamers.
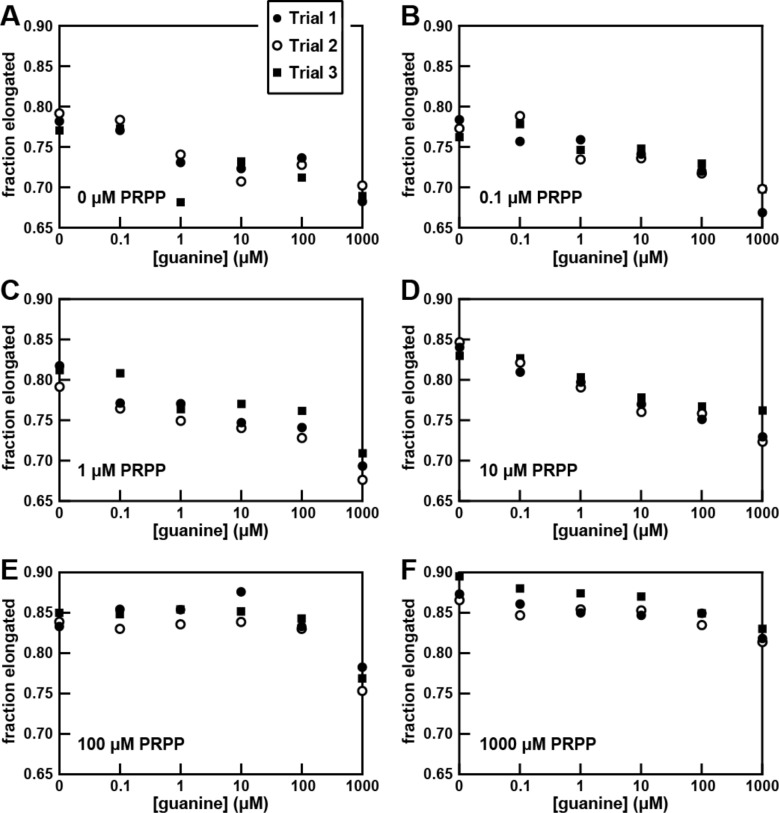
(A) Sequence and secondary structure of the tandem guanine-PRPP riboswitch derived from the codA gene of B. megaterium. An alternative RNA structure is depicted to include the terminator stem followed by a U-rich tract, which forms based on the relative abundance of each ligand. (B) PAGE analyses of single-round transcription termination assays of WT and various mutant B. megaterium codA tandem riboswitches in the presence or absence of the two ligands. FL and T denote full length and terminated transcripts, respectively. (C) Reporter gene expression of B. subtilis containing wild-type (WT) and mutant (M3 through M5) B. megaterium tandem riboswitch-lacZ reporter fusion constructs as described in (A). Cells were grown in rich (lysogeny broth) medium or glucose minimal medium (GMM) containing either no (–) or 10 µM additional guanine (gua) and 50 µg mL-1 X-gal. (D) Plot of the fraction of full length B. megaterium codA tandem riboswitch transcripts contributing to the total number of transcripts as a function of both guanine and PRPP concentrations. Data shown are the average of three experiments (see Figure 5—figure supplement 1).