Figure 2.
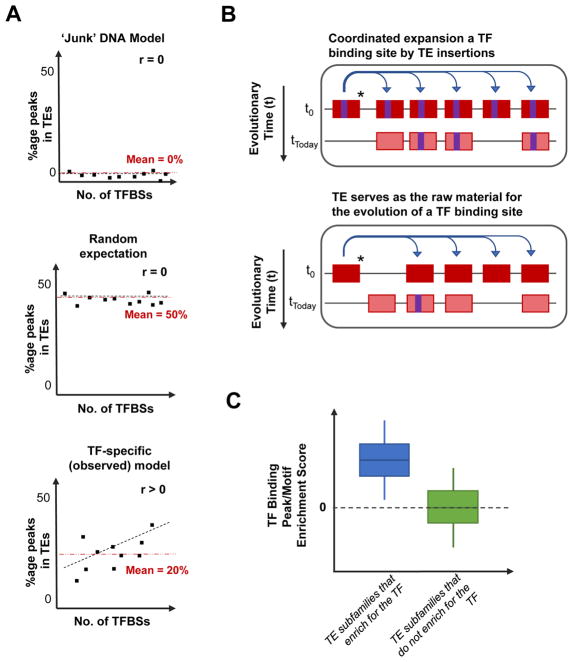
Model for understanding the evolution of TF binding sites in TEs. A: Setting the expectation for how many TF binding sites exist in TEs. Looking at various TFs (each represented by a dot in the panels), under the model of TEs being ‘junk’ DNA (upper panel), the expectation is that all TEs will have no TF binding sites because TEs are non-functional sequences. Alternatively, another common expectation is that TEs constitute almost half of the human genome sequence, therefore by chance, 50% of all TF binding sites will occur in TEs (centre panel; labelled “Random Expectation”). However, the observation is quite contrary to the previous two models (bottom panel; labelled “TF-specific (observed)). We have observed that the percentage of TF binding sites occurring in TEs ranges from 2% to 40% (average: 20%; red dotted-line in the panel) in both human and mouse cell types. It is noteworthy that the percentage of TF binding sites in TEs positively correlates with the number of TF binding sites in the genome. B: Co-ordinated expansion of TE subfamilies causing an increase in the number of TE-derived TF binding sites in the genome (upper panel;), versus TEs evolving TF binding sites by neutral evolution (lower panel). In the two models, a TE (red rectangle) capable of transposing (*) is shown to distribute across the genome. A TE with a TF binding sites (purple rectangle) will spread the TF binding site along with the TE as it transposes (upper panel). Alternatively, a TE without a binding site could serve as raw material for the evolution of new TF binding sites, based on mutations acquired (lower panel). C: Schematic representation of the enrichment scores of TEs belonging to a subfamily that enriches for a TF’s binding site (ChIP-seq peaks of sequence motifs), or TEs belonging to a subfamily that does not enrich for TF binding sites.