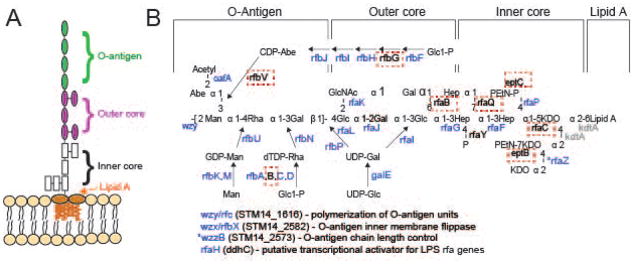
Figure 2. Mutants in the LPS biosynthetic pathway are strongly selected for during P22 infection.
A) The S. enterica serovar Typhimurium O-antigen is shown diagrammatically and in more detail in B), along with pathways of synthesis of the sugars in its trimer repeat unit (shown in square brackets) (reviewed by (Raetz et al., 1996)). Genes, in which mutants slowed P22 infection as measured by the Tn5 experiment, are indicated in blue at the bonds these enzymes catalyze. Genes in which mutation had no significant effects on P22 infection are indicated in black text, and highlighted with a red dashed box. Genes for which mutants were not assayed because the gene is essential or near essential for survival of the host in rich media are indicated in gray. LPS synthesis proceeds generally from right to left in the figure; Wzy/Rfc protein adds tetrasaccharide units to the growing O-antigen chain; RfbP protein adds the first galactose of the trisaccharide repeat to bactoprenol and RfaL transfers the O-antigen chain from there to LPS core-lipid A. Abbreviations are as follows: Glc, glucose; Rha, rhamnose; GlcNAc, N-acetylglucosamine; Man, mannose; OAc, O-acetyl; Hep, heptose; Gal, galactose; KDO, 2-keto-3-deoxyoctulosonic acid; P, phosphate, PEtN, phosphoethanolamine.