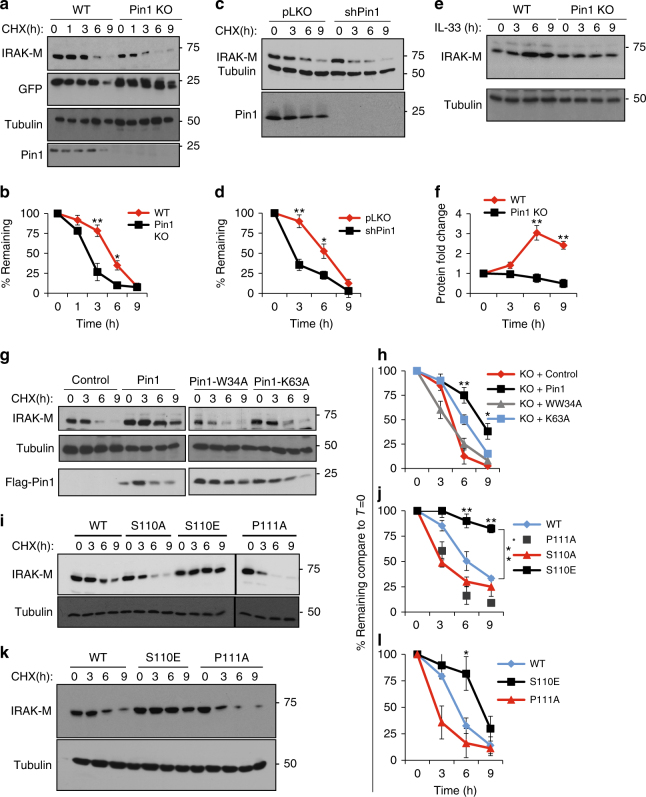
Fig. 4.
PIN1 increases IRAK-M protein stabilization. a After IRAK-M and GFP were coexpressed in WT and PIN1 KO MEFs for 24 h, cells were split equally into 5 dishes. 24 h later cells were treated with cyclohexamide and harvested to monitor IRAK-M, GFP and PIN1 levels at the time points indicated. b Quantification of 3 independent experiments as in a. c WT MEFs stably expressing the TET on inducible short hairpin for PIN1 (shPin1) or pLKO as a control, were expressed with IRAK-M, followed by induction with Doxycycline for 18 h prior to the cyclohexamide chase. d Quantification of 3 independent experiments as in c. e BMDCs from WT or PIN1 KO mice were treated with IL-33 and the levels of IRAK-M were monitored at different time points after induction. f Quantification of 3 independent experiments as in e. g PIN1 KO MEFs were expressed with IRAK-M alone or in combination with PIN1 or its mutants W34A or K63A for 24 h, followed by the cyclohexamide chase to assay IRAK-M stability. h Quantification of 3 independent experiments as in g. i IRAK-M or its different mutants; S110A, S110E and P111A were expressed in WT MEFs, followed by the cyclohexamide chase to assay IRAK-M stability. j Quantification of 3 independent experiments as in i. k IRAK-M or its different mutants were stably expressed in DC2.4 cells, followed by the cyclohexamide chase to monitor IRAK-M stability. l Quantification of 3 independent experiments as in k. The data were analyzed by a Student’s two-tailed t test and the values are reported as mean ± standard errors of the means (SEM). *- statistical significance (P < 0.05), **- significance (P < 0.01)