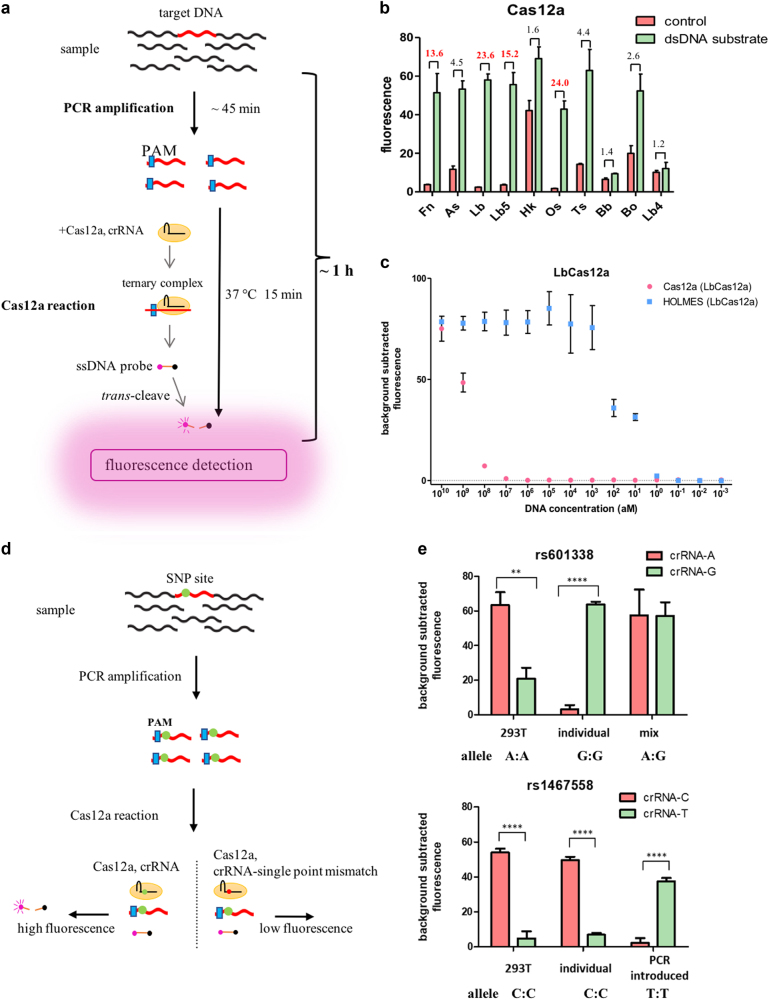
Fig. 1. HOLMES is a rapid, simple and efficient method for nucleic acid detection.
a An illustration of HOLMES. To detect a target DNA, specific amplification of the target DNA by either PCR or other isothermal amplification methods will be performed, and a crRNA guide sequence is specially designed, targeting a region in the target DNA. The PAM sequence can be designed on the primers and introduced during amplification. After that, the amplicon was mixed with the Cas12a/crRNA complex, and a ternary complex forms if the target DNA exists. Upon the formation of the ternary complex, the quenched fluorescent ssDNA reporter is trans-cleaved, illuminating the fluorescence. b Comparison of the signal-to-noise values of trans-cleavage by ten Cas12a proteins from different species. The reaction system included Cas12a, crRNA (crRNA-T1), target DNA (pUC18-T1) and quenched fluorescent ssDNA (HEX-N12-BHQ1), and the target DNA was omitted in the negative control. The signal-to-noise values were labeled and values larger than 10 were shown in red (n = 3 technical replicates; bars represent the mean ± SEM). Fn Francisella tularensis; As Acidaminococcus sp. BV3L6; Lb Lachnospiraceae bacterium ND2006; Lb5 Lachnospiraceae bacterium NC2008; HK Helcococcus kunzii ATCC 51366; Os Oribacterium sp. NK2B42; Ts Thiomicrospira sp. XS5; Bb Bacteroidales bacterium KA00251; Bo Bacteroidetes oral taxon 274 str. F0058; Lb4 Lachnospiraceae bacterium MC2017. c Detection sensitivity of Cas12a alone or Cas12a combined with PCR amplification (i.e., HOLMES). Serially diluted pUC18-T1 plasmid was employed as the target dsDNA. (n = 3 technical replicates; bars represent the mean ± SEM). d Schematic of human SNP genotyping by HOLMES. The amplification of a target DNA containing the SNP locus is almost the same as described in Fig. 1a, and design of primers and introduction of the PAM site are detailed in Supplementary Figure S3. To detect an SNP, more than one crRNA is needed, targeting different genotypes. e HOLMES correctly genotyped different human SNP loci in HEK293T, a candidate individual, and the PCR-generated templates (n = 3 technical replicates; two-tailed Student’s t-test; **p < 0.01; ****p < 0.0001; bars represent the mean ± SEM). Genotypes verified by Sanger sequencing were annotated below each plot, and the results of other SNP loci could be found in Supplementary Figure S4a