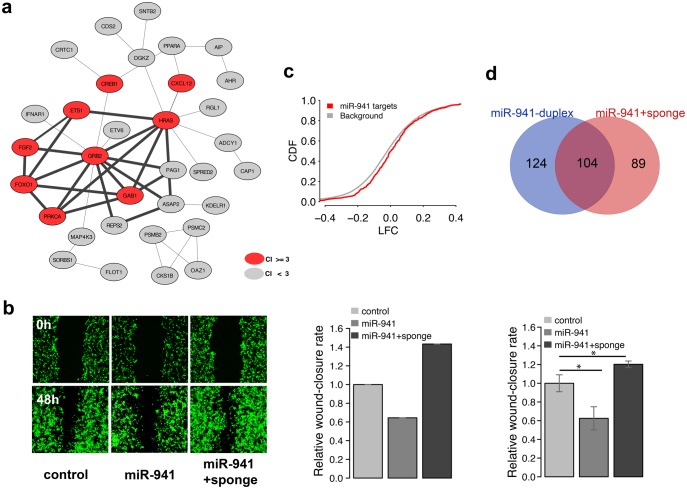
Fig. 6.
TP73-AS1 expression blocks miR-941-driven migration inhibition. (a) The bold edges indicate protein–protein interaction network module constructed based on experimentally verified miR-941 targets. The red nodes indicate genes linked to cell migration terms by publications: citation index (CI) ≥ 3. (b) The microscope images of cell density in a wound healing assay (left) and quantified wound efficiency (middle and right) after transient transfection with empty SD1266 vector (control), vector carrying miR-941 cluster region containing seven miR-941 precursors (miR-941), and vector carrying both miR-941 cluster region and TP73-AS1 sponge region (miR-941 + sponge). The middle and right panels represent the results of two independent transfection experiments, the right one conducted in triplicates. The significance of wound closure difference estimated using a t-test is shown above the bars: * P < 0.05. (c) Cumulative distribution of log2-transformed gene expression fold-change (LFC) values for genes containing predicted miR-941 target sites (red) and all other expressed genes (grey) after transfection with control and miR941 + sponge vectors into 293T cells. The y-axis shows the cumulative distribution function (CDF) of LFC distribution. (d) The overlap between miR-941 target genes (purple) identified in miR-941 duplex transfection experiment (blue) and miR941 + sponge vector transfection experiment (red).