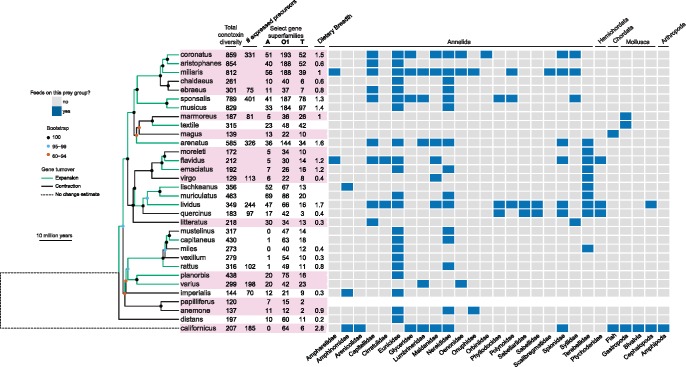
Fig. 5.
Diet and conotoxin evolution in a phylogenetic context. Time-calibrated maximum likelihood phylogeny of 32 Conidae species generated from concatenated alignment of 4,441 exons. Phylogeny is rooted with Californiconus californicus. Branches are colored based on net gains or losses in total conotoxin diversity based on CAFE v3.1 analyses. Recognized subgenera are alternately colored pink. Total conotoxin diversity, the number of expressed precursors for species with transcriptomes, size estimates for commonly studied gene superfamilies, and dietary breadth displayed next to tip names. Recorded observations of each species preying on each of the 27 represented prey families shown in the matrix adjacent to the phylogeny, with cells colored based on whether or not a species has been observed to feed on that prey family (gray = no, blue = yes). Phylum level classifications are shown at the top of the diet matrix and family level classifications are shown at the bottom of the diet matrix.