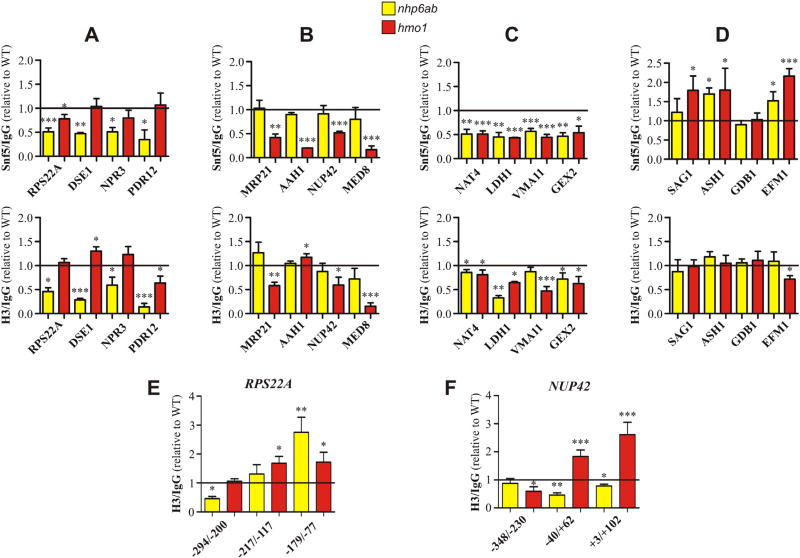
Fig. 3.
Histone H3 occupancy is affected at gene promoters undergoing reduction of SWI/SNF occupancy upon deletion of NHP6A/B and/or deletion of HMO1. ChIP-qPCR analyses were performed to determine changes in Snf5 and H3 occupancy generated by deletion of NHP6A/B (yellow bars) or HMO1 (red bars) for selected genes listed in Table 1. The groups of genes depicted in figures A, B, C, and D are in correspondence with the groups of genes listed in Table 1. Values obtained from the qPCR reactions for all genes in wild-type and deletion mutant strains were expressed as “times over IgG” and the resulting values obtained for the deletion mutants are expressed in the graphs relative to the corresponding values obtained for the wild-type strain. Thus, the horizontal black line is representative of occupancy levels in the wild-type strain. Data in each graph correspond to an assay representative of two independent assays, each performed in triplicate. Error bars represent one standard deviation. Asterisks denote a statistically significant difference (*p < 0.05; **p < 0.01; ***p < 0.001), as deducted from the t-test. Top (figures A–D): ChIP-qPCR analysis for Snf5. Bottom (figures A–D): ChIP-qPCR analysis for histone H3. Positions given here and in the figures are relative to the translation start site. Supplementary Fig. S8 is an expanded version of figures A–D. A) Analysis for genes undergoing reduction of SWI/SNF occupancy at their promoter/TSS upon deletion of NHP6A/B, as determined by the ChIP-chip assays. Regions spanned by PCR reactions: RPS22A, −294/−200; DSE1, −97/+4; NPR3, −222/−97; PDR12, −356/−227. B) Analysis for genes undergoing reduction of SWI/SNF occupancy at their promoter/TSS upon deletion of HMO1, as determined by the ChIP-chip assays. Regions spanned by PCR reactions: MRP21, −115/−15; AAH1, −213/−119; NUP42, −348/−230; MED8, −175/−44. C) Analysis for genes undergoing reduction of SWI/SNF occupancy at their promoter/TSS in both deletion mutants, as determined by the ChIP-chip assays. Regions spanned by PCR reactions: NAT4, −162/−62; LDH1, −94/+8; VMA11, −210/−96; GEX2, −356/−223. D) Analysis for genes where deletion of neither NHP6A/B nor HMO1 affects SWI/SNF binding to the promoter/TSS, as determined by the ChIP-chip assays. Regions spanned by PCR reactions: SAG1, −101/+2; ASH1, −145/−43; GDB1, −201/−73; EFM1, −226/−115. E) ChIP-qPCR analysis of histone H3 occupancy at different stretches of the RPS22A promoter. F) ChIP-qPCR analysis of histone H3 occupancy at different stretches of the NUP42 promoter.