FIG 6.
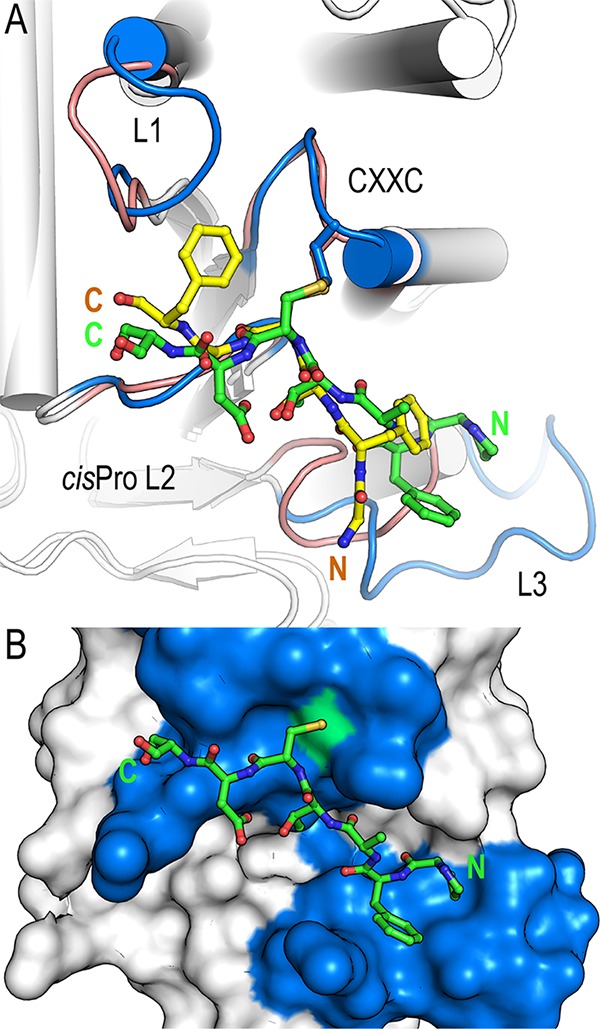
Comparison of BpsDsbA in complex with GFSCGFBpspep and E. coli DsbA in complex with the E. coli DsbB-derived heptamer peptide PFATCDSEcpep. (A) Comparison of EcDsbA-PFATCDS (PDB accession number 4TKY [24]) and BpsDsbA-GFSCGF. The crystal structure of each complex is shown in cartoon representation, with helices represented by cylinders. Active-site loops are labeled and highlighted for EcDsbA (blue) and BpsDsbA (salmon), and the respective bound peptides are shown as sticks (PFATCDSEcpep in green and GFSCGFBpspep in yellow). The C-terminal sections of GFSCGFBpspep and PFATCDSEcpep superimpose closely, and the cysteine of each peptide and adjacent main-chain atoms superimpose almost exactly. In contrast, the two peptides deviate significantly at their N termini, with a distance of up to 5 Å between the C-α of the equivalent residues 99 GlyBpspep and 101 PheEcpep. This deviation is a consequence of the shorter loop L3 in BpsDsbA than in EcDsbA. (B) In EcDsbA, a longer L3 loop forms an extended groove immediately beneath the active site to accommodate the N-terminal proline and phenylalanine. EcDsbA is shown in surface representation. Coloring is as described in the legend to panel A. The active-site cysteine (EcDsbA Cys 33) is highlighted in green. The N and C termini of the peptides are labeled in each panel.
