Fig. 6.
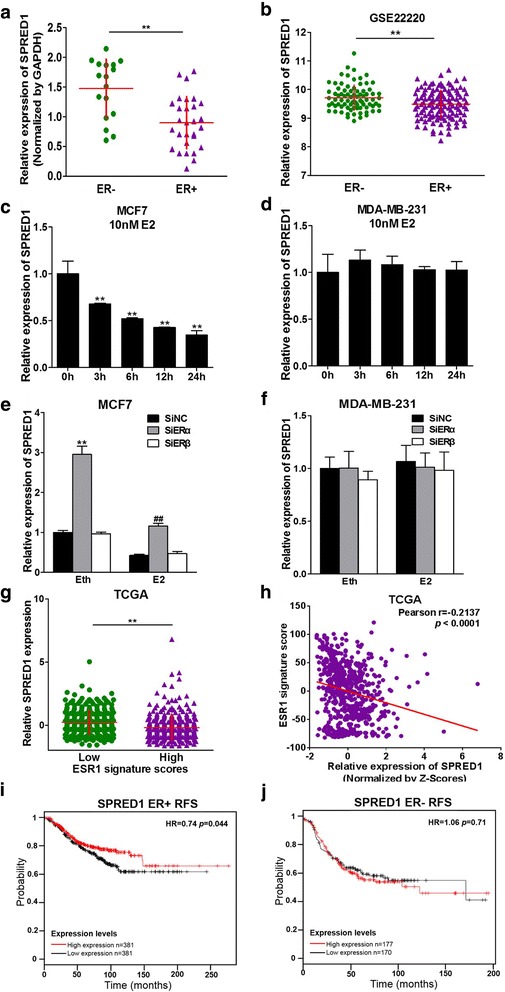
SPRED1 is down-regulated by E2 and negatively correlated to ER status in BC cells. a The expression levels of SPRED1 in 46 ER+ and ER- BC tissues were analyzed by qRT-PCR and normalized to GAPDH expression levels. ** indicates significant difference compared to normal tissues at P < 0.01. b GEO2R dataset GSE22220 was used to analyze the expression levels of SPRED1 in ER-negative or ER-positive tissues. ** indicates significant difference between ER-negative and ER-positive tumors at P < 0.01. c, d ER+ BC cells MCF7 and ER- BC cells MDA-MB-231 were cultured with estrogen-free medium for 72 h, then treated with 10 nM E2 or equal amount of solvent Eth. The expression levels of SPRED1 were analyzed using qRT-PCR, and normalized to the values of the Eth control. Data were presented as the means ± SD from three independent experiments with triple replicates per experiment. ** indicates significant difference compared to the 0 h group at P < 0.01. e, f Cells were treated as above, then transfected with 50 nM ERα siRNAs, ERβ siRNAs or negative control siRNAs (siNC). After 24 h, the cells were treated with 10 nM E2 or Eth, the expression levels of SPRED1 were analyzed using qRT-PCR, and normalized to the value of siNC+Eth group. ** indicates significant difference between siNC+Eth and siERα+Eth group at P < 0.01. ## indicates significant difference between siNC+E2 and siERα+E2 group at P < 0.01. g Primary human breast cancers of TCGA BRCA dataset were classified to high or low ESR1 signature score group, and the expression levels of SPRED1 were analyzed in these two groups. The details were described in methods. ** indicates significant difference between low ESR1 signature score group and high ESR1 signature group. h Pearson’s correlation analysis was used to determine the correlation between the expression levels of ESR1 signature score and SPRED1 expression levels. i, j The Kaplan Meier plotter was used to detect the relapse free survival (RFS) of SPRED1 in ER+ and ER- BC patients, respectively
